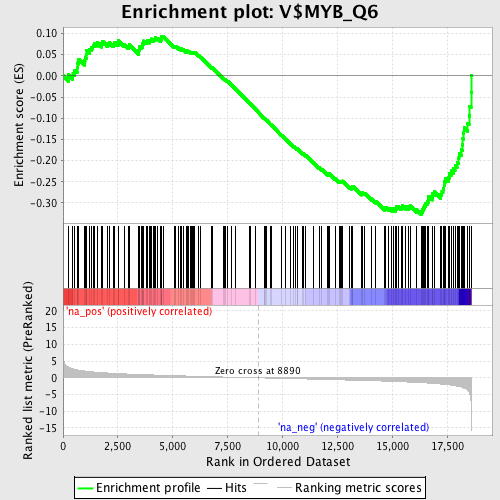
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MYB_Q6 |
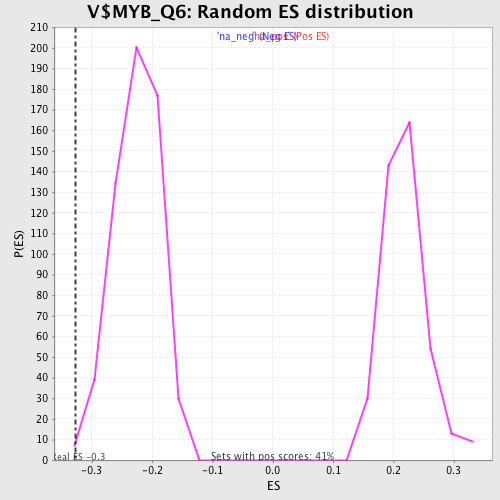
| Enrichment Score (ES) | -0.32731685 |
| Normalized Enrichment Score (NES) | -1.4535694 |
| Nominal p-value | 0.00681431 |
| FDR q-value | 0.5037867 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GCNT2 | 241 | 3.188 | 0.0030 | No | ||
| 2 | KLF5 | 449 | 2.682 | 0.0053 | No | ||
| 3 | PIP5K2B | 530 | 2.542 | 0.0138 | No | ||
| 4 | CNTF | 637 | 2.405 | 0.0201 | No | ||
| 5 | CDK2 | 668 | 2.362 | 0.0304 | No | ||
| 6 | HHEX | 716 | 2.284 | 0.0394 | No | ||
| 7 | P2RY2 | 975 | 2.031 | 0.0356 | No | ||
| 8 | ALDOA | 998 | 2.006 | 0.0445 | No | ||
| 9 | ANK2 | 1054 | 1.967 | 0.0515 | No | ||
| 10 | CD5 | 1061 | 1.960 | 0.0610 | No | ||
| 11 | ADRBK1 | 1212 | 1.855 | 0.0622 | No | ||
| 12 | TLE3 | 1277 | 1.821 | 0.0679 | No | ||
| 13 | SLC39A4 | 1362 | 1.760 | 0.0723 | No | ||
| 14 | MDM1 | 1439 | 1.713 | 0.0768 | No | ||
| 15 | CD44 | 1560 | 1.647 | 0.0786 | No | ||
| 16 | ODF2 | 1757 | 1.548 | 0.0757 | No | ||
| 17 | NEDD8 | 1796 | 1.534 | 0.0814 | No | ||
| 18 | RHOV | 2021 | 1.443 | 0.0766 | No | ||
| 19 | GCAT | 2101 | 1.411 | 0.0794 | No | ||
| 20 | SDC1 | 2306 | 1.334 | 0.0750 | No | ||
| 21 | CDCA3 | 2352 | 1.319 | 0.0793 | No | ||
| 22 | CAMK2A | 2506 | 1.273 | 0.0774 | No | ||
| 23 | GTF3C2 | 2521 | 1.268 | 0.0830 | No | ||
| 24 | SLC16A6 | 2781 | 1.188 | 0.0750 | No | ||
| 25 | MYO18A | 2973 | 1.133 | 0.0703 | No | ||
| 26 | FBXL2 | 3019 | 1.117 | 0.0735 | No | ||
| 27 | BAP1 | 3440 | 1.008 | 0.0558 | No | ||
| 28 | CITED2 | 3443 | 1.007 | 0.0608 | No | ||
| 29 | CLDN8 | 3467 | 0.999 | 0.0646 | No | ||
| 30 | FSTL5 | 3489 | 0.993 | 0.0684 | No | ||
| 31 | ANKRD17 | 3573 | 0.969 | 0.0688 | No | ||
| 32 | NKX2-8 | 3623 | 0.957 | 0.0710 | No | ||
| 33 | MAP4K4 | 3624 | 0.957 | 0.0758 | No | ||
| 34 | CHRDL1 | 3674 | 0.945 | 0.0779 | No | ||
| 35 | HOXC6 | 3681 | 0.944 | 0.0824 | No | ||
| 36 | THRAP1 | 3795 | 0.918 | 0.0809 | No | ||
| 37 | CDH13 | 3823 | 0.911 | 0.0840 | No | ||
| 38 | FLI1 | 3945 | 0.885 | 0.0819 | No | ||
| 39 | PCTK2 | 3980 | 0.879 | 0.0845 | No | ||
| 40 | ZYX | 4014 | 0.872 | 0.0871 | No | ||
| 41 | DSCR1L1 | 4132 | 0.844 | 0.0850 | No | ||
| 42 | ASPA | 4181 | 0.832 | 0.0866 | No | ||
| 43 | HOXB3 | 4192 | 0.829 | 0.0902 | No | ||
| 44 | STK11IP | 4296 | 0.799 | 0.0887 | No | ||
| 45 | C6 | 4442 | 0.771 | 0.0847 | No | ||
| 46 | PFN2 | 4464 | 0.764 | 0.0874 | No | ||
| 47 | TRPC1 | 4477 | 0.762 | 0.0906 | No | ||
| 48 | NEUROD6 | 4499 | 0.756 | 0.0933 | No | ||
| 49 | BHLHB5 | 4559 | 0.747 | 0.0938 | No | ||
| 50 | IGSF1 | 5068 | 0.645 | 0.0695 | No | ||
| 51 | GNG3 | 5123 | 0.634 | 0.0698 | No | ||
| 52 | CACNA1G | 5250 | 0.611 | 0.0661 | No | ||
| 53 | IGFBP4 | 5346 | 0.592 | 0.0639 | No | ||
| 54 | FUT11 | 5405 | 0.582 | 0.0637 | No | ||
| 55 | WFS1 | 5481 | 0.568 | 0.0625 | No | ||
| 56 | FBXO24 | 5637 | 0.540 | 0.0568 | No | ||
| 57 | RAB4B | 5647 | 0.539 | 0.0590 | No | ||
| 58 | MNT | 5709 | 0.527 | 0.0584 | No | ||
| 59 | UBE2S | 5798 | 0.509 | 0.0562 | No | ||
| 60 | KCNK7 | 5862 | 0.495 | 0.0553 | No | ||
| 61 | RHOC | 5919 | 0.487 | 0.0547 | No | ||
| 62 | TTC16 | 5964 | 0.481 | 0.0547 | No | ||
| 63 | PHF7 | 5982 | 0.476 | 0.0562 | No | ||
| 64 | RAB3C | 6175 | 0.441 | 0.0480 | No | ||
| 65 | ARHGEF6 | 6282 | 0.422 | 0.0444 | No | ||
| 66 | STC1 | 6780 | 0.343 | 0.0192 | No | ||
| 67 | GMPR2 | 6808 | 0.338 | 0.0194 | No | ||
| 68 | RFX4 | 7291 | 0.252 | -0.0054 | No | ||
| 69 | OTX1 | 7373 | 0.238 | -0.0086 | No | ||
| 70 | ONECUT1 | 7392 | 0.235 | -0.0084 | No | ||
| 71 | BSCL2 | 7485 | 0.221 | -0.0123 | No | ||
| 72 | ALOX12B | 7696 | 0.189 | -0.0227 | No | ||
| 73 | SILV | 7856 | 0.164 | -0.0305 | No | ||
| 74 | B3GALT2 | 8511 | 0.060 | -0.0656 | No | ||
| 75 | CAPZA1 | 8529 | 0.058 | -0.0662 | No | ||
| 76 | MB | 8774 | 0.021 | -0.0794 | No | ||
| 77 | TNNI1 | 9158 | -0.040 | -0.0999 | No | ||
| 78 | STC2 | 9167 | -0.041 | -0.1001 | No | ||
| 79 | JARID2 | 9203 | -0.046 | -0.1018 | No | ||
| 80 | SLC9A7 | 9284 | -0.059 | -0.1058 | No | ||
| 81 | USP5 | 9438 | -0.082 | -0.1137 | No | ||
| 82 | NOTCH2 | 9499 | -0.089 | -0.1165 | No | ||
| 83 | PFTK1 | 9946 | -0.159 | -0.1399 | No | ||
| 84 | SPOCK2 | 10151 | -0.188 | -0.1500 | No | ||
| 85 | YWHAE | 10376 | -0.220 | -0.1610 | No | ||
| 86 | HOXB4 | 10509 | -0.243 | -0.1669 | No | ||
| 87 | FMOD | 10604 | -0.260 | -0.1707 | No | ||
| 88 | BDNF | 10661 | -0.271 | -0.1724 | No | ||
| 89 | PHC1 | 10676 | -0.273 | -0.1718 | No | ||
| 90 | PRICKLE1 | 10918 | -0.311 | -0.1833 | No | ||
| 91 | HOXB8 | 10969 | -0.320 | -0.1844 | No | ||
| 92 | GNAT1 | 11065 | -0.337 | -0.1878 | No | ||
| 93 | NXPH1 | 11433 | -0.398 | -0.2057 | No | ||
| 94 | LNPEP | 11667 | -0.435 | -0.2161 | No | ||
| 95 | LRFN5 | 11798 | -0.455 | -0.2209 | No | ||
| 96 | TYSND1 | 12052 | -0.495 | -0.2321 | No | ||
| 97 | HOXB5 | 12082 | -0.501 | -0.2311 | No | ||
| 98 | HOXA3 | 12126 | -0.508 | -0.2309 | No | ||
| 99 | IVL | 12403 | -0.556 | -0.2431 | No | ||
| 100 | RAPSN | 12581 | -0.592 | -0.2497 | No | ||
| 101 | SNRPD1 | 12627 | -0.601 | -0.2491 | No | ||
| 102 | JPH3 | 12680 | -0.609 | -0.2488 | No | ||
| 103 | FGF12 | 12727 | -0.617 | -0.2482 | No | ||
| 104 | RGN | 13054 | -0.678 | -0.2625 | No | ||
| 105 | RARG | 13146 | -0.690 | -0.2639 | No | ||
| 106 | NR3C2 | 13175 | -0.694 | -0.2619 | No | ||
| 107 | HNRPF | 13209 | -0.699 | -0.2602 | No | ||
| 108 | PCSK1N | 13601 | -0.769 | -0.2775 | No | ||
| 109 | PEX16 | 13646 | -0.777 | -0.2760 | No | ||
| 110 | PAFAH1B1 | 13732 | -0.792 | -0.2766 | No | ||
| 111 | LRCH4 | 14054 | -0.853 | -0.2897 | No | ||
| 112 | NR6A1 | 14251 | -0.893 | -0.2958 | No | ||
| 113 | CHN1 | 14646 | -0.978 | -0.3122 | No | ||
| 114 | RHOA | 14709 | -0.991 | -0.3106 | No | ||
| 115 | ADAM2 | 14839 | -1.025 | -0.3124 | No | ||
| 116 | TPP2 | 14966 | -1.049 | -0.3140 | No | ||
| 117 | ST7L | 15064 | -1.074 | -0.3138 | No | ||
| 118 | MSN | 15168 | -1.102 | -0.3138 | No | ||
| 119 | ODF4 | 15186 | -1.105 | -0.3092 | No | ||
| 120 | SSBP2 | 15279 | -1.130 | -0.3085 | No | ||
| 121 | MAMDC1 | 15432 | -1.170 | -0.3108 | No | ||
| 122 | TGM5 | 15465 | -1.179 | -0.3066 | No | ||
| 123 | DDR2 | 15614 | -1.216 | -0.3085 | No | ||
| 124 | ARHGAP12 | 15742 | -1.249 | -0.3091 | No | ||
| 125 | FRAS1 | 15822 | -1.274 | -0.3070 | No | ||
| 126 | REPS1 | 16102 | -1.359 | -0.3152 | No | ||
| 127 | MBD3 | 16326 | -1.438 | -0.3201 | Yes | ||
| 128 | AK2 | 16372 | -1.453 | -0.3152 | Yes | ||
| 129 | SPAG9 | 16434 | -1.479 | -0.3110 | Yes | ||
| 130 | FBXL14 | 16463 | -1.492 | -0.3050 | Yes | ||
| 131 | DACT1 | 16532 | -1.515 | -0.3011 | Yes | ||
| 132 | FGF16 | 16598 | -1.541 | -0.2969 | Yes | ||
| 133 | GRIN2A | 16636 | -1.558 | -0.2910 | Yes | ||
| 134 | EIF4G2 | 16666 | -1.569 | -0.2847 | Yes | ||
| 135 | PREB | 16823 | -1.639 | -0.2849 | Yes | ||
| 136 | TCTA | 16850 | -1.650 | -0.2780 | Yes | ||
| 137 | PCDH9 | 16924 | -1.682 | -0.2734 | Yes | ||
| 138 | ESRRG | 17214 | -1.841 | -0.2798 | Yes | ||
| 139 | HNRPUL1 | 17266 | -1.878 | -0.2731 | Yes | ||
| 140 | YPEL1 | 17315 | -1.905 | -0.2661 | Yes | ||
| 141 | CSAD | 17360 | -1.927 | -0.2588 | Yes | ||
| 142 | SESN3 | 17381 | -1.940 | -0.2501 | Yes | ||
| 143 | FYN | 17408 | -1.958 | -0.2417 | Yes | ||
| 144 | LTBP1 | 17570 | -2.081 | -0.2399 | Yes | ||
| 145 | DLX1 | 17598 | -2.107 | -0.2307 | Yes | ||
| 146 | UBE2H | 17683 | -2.162 | -0.2244 | Yes | ||
| 147 | ING3 | 17779 | -2.247 | -0.2182 | Yes | ||
| 148 | RIN1 | 17863 | -2.343 | -0.2109 | Yes | ||
| 149 | PABPC1 | 17954 | -2.454 | -0.2034 | Yes | ||
| 150 | SEMA7A | 18015 | -2.541 | -0.1939 | Yes | ||
| 151 | UGCGL1 | 18044 | -2.574 | -0.1824 | Yes | ||
| 152 | UTX | 18144 | -2.747 | -0.1740 | Yes | ||
| 153 | HES6 | 18205 | -2.877 | -0.1627 | Yes | ||
| 154 | TENC1 | 18221 | -2.910 | -0.1489 | Yes | ||
| 155 | CACNB2 | 18234 | -2.938 | -0.1347 | Yes | ||
| 156 | EDG5 | 18271 | -3.047 | -0.1213 | Yes | ||
| 157 | MGLL | 18444 | -3.687 | -0.1121 | Yes | ||
| 158 | GRIA3 | 18502 | -4.254 | -0.0937 | Yes | ||
| 159 | GNAS | 18538 | -4.797 | -0.0715 | Yes | ||
| 160 | SOCS2 | 18593 | -6.945 | -0.0394 | Yes | ||
| 161 | PTPN22 | 18603 | -8.056 | 0.0007 | Yes |