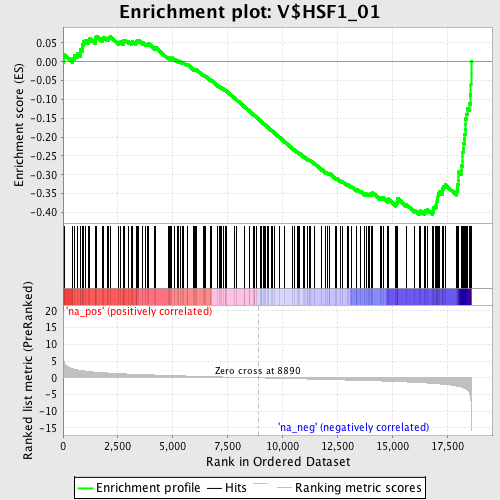
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$HSF1_01 |
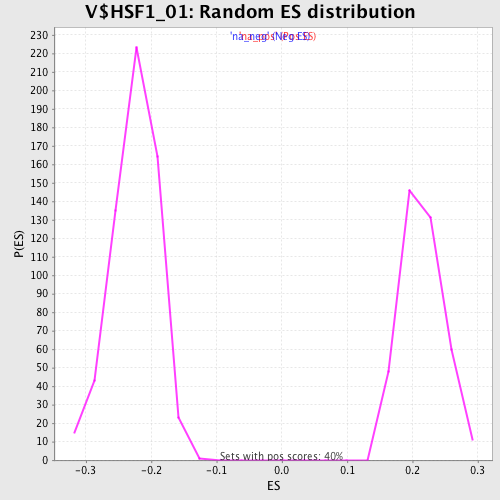
| Enrichment Score (ES) | -0.40522322 |
| Normalized Enrichment Score (NES) | -1.8030789 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.24542841 |
| FWER p-Value | 0.219 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IGFBP5 | 40 | 4.986 | 0.0197 | No | ||
| 2 | RAB6B | 443 | 2.692 | 0.0098 | No | ||
| 3 | ADAMTS2 | 511 | 2.571 | 0.0174 | No | ||
| 4 | CNTF | 637 | 2.405 | 0.0212 | No | ||
| 5 | ADCY6 | 789 | 2.199 | 0.0227 | No | ||
| 6 | MAPK14 | 804 | 2.181 | 0.0315 | No | ||
| 7 | BCL6 | 880 | 2.112 | 0.0368 | No | ||
| 8 | CYLD | 888 | 2.101 | 0.0456 | No | ||
| 9 | RASGRP2 | 920 | 2.073 | 0.0530 | No | ||
| 10 | SUHW3 | 1010 | 1.999 | 0.0570 | No | ||
| 11 | GAD2 | 1154 | 1.897 | 0.0576 | No | ||
| 12 | ABCD1 | 1221 | 1.848 | 0.0621 | No | ||
| 13 | DNAJB1 | 1468 | 1.694 | 0.0562 | No | ||
| 14 | LENEP | 1475 | 1.691 | 0.0633 | No | ||
| 15 | MOBP | 1526 | 1.662 | 0.0679 | No | ||
| 16 | OTX2 | 1779 | 1.542 | 0.0610 | No | ||
| 17 | ITGA8 | 1823 | 1.524 | 0.0654 | No | ||
| 18 | LUZP1 | 2009 | 1.449 | 0.0617 | No | ||
| 19 | MBNL2 | 2088 | 1.415 | 0.0637 | No | ||
| 20 | DAZL | 2151 | 1.397 | 0.0665 | No | ||
| 21 | LHFPL2 | 2545 | 1.259 | 0.0507 | No | ||
| 22 | FHL1 | 2594 | 1.244 | 0.0536 | No | ||
| 23 | PLEKHB1 | 2757 | 1.193 | 0.0500 | No | ||
| 24 | BNC2 | 2759 | 1.193 | 0.0552 | No | ||
| 25 | OTP | 2813 | 1.179 | 0.0575 | No | ||
| 26 | TRERF1 | 2988 | 1.128 | 0.0531 | No | ||
| 27 | ZFX | 3115 | 1.093 | 0.0510 | No | ||
| 28 | RPH3AL | 3168 | 1.080 | 0.0530 | No | ||
| 29 | HIPK1 | 3322 | 1.040 | 0.0492 | No | ||
| 30 | GREB1 | 3340 | 1.035 | 0.0529 | No | ||
| 31 | PIP5K1A | 3378 | 1.025 | 0.0554 | No | ||
| 32 | MYOCD | 3421 | 1.014 | 0.0575 | No | ||
| 33 | FOXD2 | 3625 | 0.957 | 0.0507 | No | ||
| 34 | SLC24A4 | 3774 | 0.921 | 0.0468 | No | ||
| 35 | ARL6IP2 | 3844 | 0.906 | 0.0470 | No | ||
| 36 | SPO11 | 3871 | 0.901 | 0.0495 | No | ||
| 37 | PTGIS | 4174 | 0.833 | 0.0368 | No | ||
| 38 | LAP3 | 4226 | 0.818 | 0.0377 | No | ||
| 39 | FSTL3 | 4783 | 0.700 | 0.0106 | No | ||
| 40 | NFATC4 | 4866 | 0.684 | 0.0091 | No | ||
| 41 | ATP6V1A | 4914 | 0.676 | 0.0096 | No | ||
| 42 | FOXB1 | 4960 | 0.666 | 0.0101 | No | ||
| 43 | SOCS5 | 5078 | 0.644 | 0.0065 | No | ||
| 44 | SREBF2 | 5214 | 0.617 | 0.0019 | No | ||
| 45 | NR4A3 | 5261 | 0.609 | 0.0021 | No | ||
| 46 | TRDN | 5343 | 0.593 | 0.0003 | No | ||
| 47 | RORB | 5451 | 0.573 | -0.0030 | No | ||
| 48 | EHD1 | 5507 | 0.563 | -0.0035 | No | ||
| 49 | JAM3 | 5652 | 0.539 | -0.0089 | No | ||
| 50 | CRISP1 | 5653 | 0.538 | -0.0066 | No | ||
| 51 | BCL9 | 5956 | 0.482 | -0.0208 | No | ||
| 52 | CCM2 | 5976 | 0.478 | -0.0197 | No | ||
| 53 | GRM1 | 6024 | 0.468 | -0.0202 | No | ||
| 54 | DNAJA1 | 6059 | 0.463 | -0.0200 | No | ||
| 55 | BMPR2 | 6381 | 0.405 | -0.0357 | No | ||
| 56 | GPR17 | 6426 | 0.396 | -0.0363 | No | ||
| 57 | TYR | 6499 | 0.384 | -0.0385 | No | ||
| 58 | PERQ1 | 6700 | 0.356 | -0.0478 | No | ||
| 59 | STC1 | 6780 | 0.343 | -0.0506 | No | ||
| 60 | NEK6 | 7047 | 0.294 | -0.0637 | No | ||
| 61 | HS6ST3 | 7114 | 0.280 | -0.0661 | No | ||
| 62 | MRPS6 | 7183 | 0.269 | -0.0686 | No | ||
| 63 | GLRA3 | 7220 | 0.263 | -0.0694 | No | ||
| 64 | RFX4 | 7291 | 0.252 | -0.0721 | No | ||
| 65 | POU4F3 | 7389 | 0.236 | -0.0763 | No | ||
| 66 | OSR2 | 7401 | 0.234 | -0.0759 | No | ||
| 67 | CHML | 7457 | 0.226 | -0.0778 | No | ||
| 68 | ISYNA1 | 7797 | 0.173 | -0.0955 | No | ||
| 69 | CLDN2 | 7908 | 0.155 | -0.1008 | No | ||
| 70 | ABHD3 | 8244 | 0.102 | -0.1185 | No | ||
| 71 | MAP2K1IP1 | 8263 | 0.101 | -0.1190 | No | ||
| 72 | RNF5 | 8478 | 0.065 | -0.1303 | No | ||
| 73 | NFATC3 | 8491 | 0.063 | -0.1307 | No | ||
| 74 | STX12 | 8699 | 0.033 | -0.1418 | No | ||
| 75 | WRN | 8705 | 0.032 | -0.1419 | No | ||
| 76 | BCAP31 | 8736 | 0.028 | -0.1434 | No | ||
| 77 | PLXNC1 | 8835 | 0.011 | -0.1487 | No | ||
| 78 | SMYD1 | 9014 | -0.018 | -0.1583 | No | ||
| 79 | ADCY8 | 9027 | -0.021 | -0.1588 | No | ||
| 80 | HSPB1 | 9035 | -0.021 | -0.1591 | No | ||
| 81 | WDR12 | 9135 | -0.037 | -0.1643 | No | ||
| 82 | PTHLH | 9180 | -0.043 | -0.1665 | No | ||
| 83 | FAP | 9210 | -0.047 | -0.1679 | No | ||
| 84 | KNTC1 | 9328 | -0.066 | -0.1739 | No | ||
| 85 | DMRT1 | 9357 | -0.071 | -0.1751 | No | ||
| 86 | H2AFZ | 9497 | -0.089 | -0.1823 | No | ||
| 87 | CACNA2D2 | 9538 | -0.095 | -0.1840 | No | ||
| 88 | WNT8B | 9646 | -0.111 | -0.1894 | No | ||
| 89 | IL2 | 9880 | -0.148 | -0.2014 | No | ||
| 90 | ADAMTS9 | 10107 | -0.181 | -0.2128 | No | ||
| 91 | PURG | 10471 | -0.236 | -0.2315 | No | ||
| 92 | NR2F1 | 10524 | -0.246 | -0.2332 | No | ||
| 93 | CHST8 | 10663 | -0.271 | -0.2395 | No | ||
| 94 | PAK1IP1 | 10720 | -0.280 | -0.2413 | No | ||
| 95 | ARHGAP6 | 10792 | -0.291 | -0.2439 | No | ||
| 96 | HOXB8 | 10969 | -0.320 | -0.2520 | No | ||
| 97 | SCG2 | 10990 | -0.325 | -0.2517 | No | ||
| 98 | KCNQ1 | 11160 | -0.353 | -0.2593 | No | ||
| 99 | ANKRD22 | 11222 | -0.364 | -0.2610 | No | ||
| 100 | RIMS2 | 11271 | -0.373 | -0.2620 | No | ||
| 101 | TREX1 | 11472 | -0.403 | -0.2710 | No | ||
| 102 | A2BP1 | 11787 | -0.453 | -0.2861 | No | ||
| 103 | PAK3 | 11980 | -0.482 | -0.2944 | No | ||
| 104 | PLXDC2 | 12037 | -0.492 | -0.2952 | No | ||
| 105 | NR2C2 | 12119 | -0.507 | -0.2974 | No | ||
| 106 | TRIB1 | 12157 | -0.514 | -0.2971 | No | ||
| 107 | AGPAT1 | 12418 | -0.558 | -0.3088 | No | ||
| 108 | SIX4 | 12459 | -0.566 | -0.3085 | No | ||
| 109 | TLK1 | 12654 | -0.604 | -0.3163 | No | ||
| 110 | YWHAG | 12725 | -0.617 | -0.3174 | No | ||
| 111 | GAD1 | 12947 | -0.657 | -0.3265 | No | ||
| 112 | DNAJB5 | 13009 | -0.670 | -0.3269 | No | ||
| 113 | E2F3 | 13138 | -0.689 | -0.3308 | No | ||
| 114 | HCN4 | 13364 | -0.724 | -0.3398 | No | ||
| 115 | NRXN3 | 13391 | -0.728 | -0.3380 | No | ||
| 116 | DBI | 13553 | -0.761 | -0.3434 | No | ||
| 117 | NNAT | 13747 | -0.795 | -0.3504 | No | ||
| 118 | PPP2R5C | 13818 | -0.807 | -0.3507 | No | ||
| 119 | NT5C3 | 13916 | -0.825 | -0.3523 | No | ||
| 120 | ALS2CR8 | 13958 | -0.833 | -0.3508 | No | ||
| 121 | FLNC | 14045 | -0.852 | -0.3518 | No | ||
| 122 | PHF6 | 14052 | -0.853 | -0.3483 | No | ||
| 123 | EIF5 | 14116 | -0.866 | -0.3479 | No | ||
| 124 | PPT2 | 14446 | -0.933 | -0.3617 | No | ||
| 125 | CHD1 | 14507 | -0.946 | -0.3608 | No | ||
| 126 | ASAH2 | 14596 | -0.967 | -0.3613 | No | ||
| 127 | CREM | 14801 | -1.016 | -0.3679 | No | ||
| 128 | DMRTB1 | 14813 | -1.018 | -0.3640 | No | ||
| 129 | NOL4 | 15162 | -1.099 | -0.3781 | No | ||
| 130 | TPM2 | 15197 | -1.110 | -0.3750 | No | ||
| 131 | ATP1B2 | 15222 | -1.118 | -0.3714 | No | ||
| 132 | PIM1 | 15228 | -1.119 | -0.3668 | No | ||
| 133 | DMRTA1 | 15251 | -1.125 | -0.3630 | No | ||
| 134 | HSPA8 | 15640 | -1.221 | -0.3787 | No | ||
| 135 | RTN1 | 16028 | -1.334 | -0.3938 | No | ||
| 136 | SRP54 | 16230 | -1.403 | -0.3986 | Yes | ||
| 137 | MAP3K7IP2 | 16296 | -1.427 | -0.3958 | Yes | ||
| 138 | ELAVL4 | 16464 | -1.492 | -0.3983 | Yes | ||
| 139 | EGR3 | 16523 | -1.514 | -0.3948 | Yes | ||
| 140 | PRKAG1 | 16600 | -1.541 | -0.3922 | Yes | ||
| 141 | ACTN2 | 16842 | -1.646 | -0.3980 | Yes | ||
| 142 | HIVEP2 | 16873 | -1.660 | -0.3923 | Yes | ||
| 143 | FOS | 16885 | -1.668 | -0.3856 | Yes | ||
| 144 | EOMES | 16961 | -1.700 | -0.3822 | Yes | ||
| 145 | DUSP1 | 17035 | -1.743 | -0.3785 | Yes | ||
| 146 | SERPINH1 | 17038 | -1.745 | -0.3709 | Yes | ||
| 147 | CHORDC1 | 17055 | -1.752 | -0.3641 | Yes | ||
| 148 | ANKRD25 | 17084 | -1.770 | -0.3578 | Yes | ||
| 149 | MORF4L2 | 17087 | -1.772 | -0.3501 | Yes | ||
| 150 | PACSIN3 | 17142 | -1.801 | -0.3452 | Yes | ||
| 151 | RBPMS | 17275 | -1.884 | -0.3440 | Yes | ||
| 152 | GRIK1 | 17291 | -1.894 | -0.3365 | Yes | ||
| 153 | PIP5K3 | 17341 | -1.918 | -0.3307 | Yes | ||
| 154 | AAMP | 17411 | -1.958 | -0.3259 | Yes | ||
| 155 | OPRD1 | 17923 | -2.412 | -0.3430 | Yes | ||
| 156 | TNFRSF19 | 17972 | -2.483 | -0.3347 | Yes | ||
| 157 | SMAD1 | 17991 | -2.512 | -0.3246 | Yes | ||
| 158 | PCDH10 | 18010 | -2.536 | -0.3145 | Yes | ||
| 159 | PDE6D | 18017 | -2.542 | -0.3036 | Yes | ||
| 160 | HSPD1 | 18018 | -2.544 | -0.2924 | Yes | ||
| 161 | ABCC5 | 18147 | -2.750 | -0.2873 | Yes | ||
| 162 | HSPH1 | 18163 | -2.779 | -0.2759 | Yes | ||
| 163 | CBX3 | 18189 | -2.840 | -0.2647 | Yes | ||
| 164 | HNRPA2B1 | 18191 | -2.844 | -0.2523 | Yes | ||
| 165 | HSPB8 | 18192 | -2.845 | -0.2398 | Yes | ||
| 166 | UBE2E2 | 18235 | -2.939 | -0.2292 | Yes | ||
| 167 | GSN | 18264 | -3.025 | -0.2174 | Yes | ||
| 168 | ST13 | 18284 | -3.075 | -0.2049 | Yes | ||
| 169 | FKBP4 | 18304 | -3.127 | -0.1922 | Yes | ||
| 170 | DMD | 18322 | -3.185 | -0.1791 | Yes | ||
| 171 | JUN | 18337 | -3.236 | -0.1656 | Yes | ||
| 172 | GATA3 | 18340 | -3.255 | -0.1514 | Yes | ||
| 173 | CCT3 | 18392 | -3.448 | -0.1390 | Yes | ||
| 174 | ELOVL5 | 18418 | -3.580 | -0.1247 | Yes | ||
| 175 | PLXNA2 | 18526 | -4.536 | -0.1105 | Yes | ||
| 176 | EPHA2 | 18572 | -5.699 | -0.0879 | Yes | ||
| 177 | CISH | 18589 | -6.631 | -0.0596 | Yes | ||
| 178 | CCNG1 | 18592 | -6.932 | -0.0293 | Yes | ||
| 179 | SOCS2 | 18593 | -6.945 | 0.0012 | Yes |