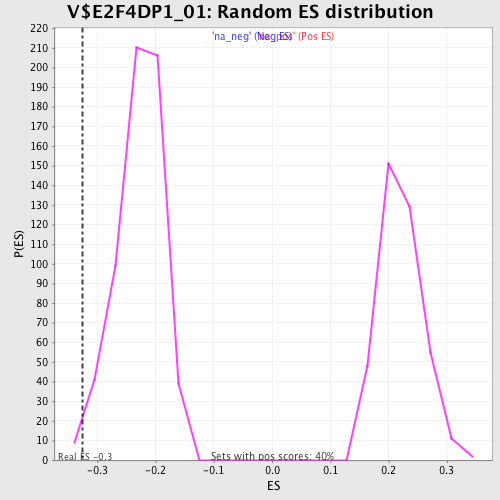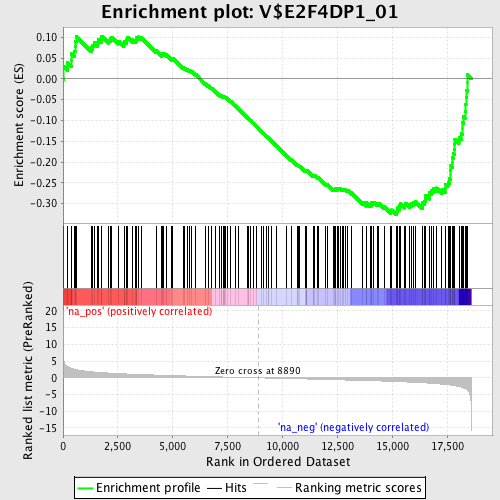
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$E2F4DP1_01 |
| Enrichment Score (ES) | -0.32601464 |
| Normalized Enrichment Score (NES) | -1.4332224 |
| Nominal p-value | 0.0066225166 |
| FDR q-value | 0.5140147 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EFNA5 | 26 | 5.882 | 0.0303 | No | ||
| 2 | MAPT | 202 | 3.363 | 0.0389 | No | ||
| 3 | CCNT1 | 369 | 2.827 | 0.0451 | No | ||
| 4 | TBX6 | 373 | 2.818 | 0.0601 | No | ||
| 5 | ADAMTS2 | 511 | 2.571 | 0.0666 | No | ||
| 6 | HIRA | 545 | 2.526 | 0.0784 | No | ||
| 7 | KLF4 | 566 | 2.494 | 0.0907 | No | ||
| 8 | SLCO3A1 | 605 | 2.432 | 0.1018 | No | ||
| 9 | TLE3 | 1277 | 1.821 | 0.0752 | No | ||
| 10 | MAT2A | 1356 | 1.763 | 0.0805 | No | ||
| 11 | FANCD2 | 1411 | 1.728 | 0.0869 | No | ||
| 12 | PRPF4B | 1567 | 1.644 | 0.0873 | No | ||
| 13 | SFRS1 | 1598 | 1.628 | 0.0945 | No | ||
| 14 | IL4I1 | 1735 | 1.557 | 0.0955 | No | ||
| 15 | PRKDC | 1753 | 1.550 | 0.1029 | No | ||
| 16 | MXD3 | 2068 | 1.420 | 0.0936 | No | ||
| 17 | LHX5 | 2156 | 1.394 | 0.0963 | No | ||
| 18 | HMGA1 | 2220 | 1.366 | 0.1003 | No | ||
| 19 | IPO7 | 2523 | 1.267 | 0.0908 | No | ||
| 20 | DNAJC9 | 2776 | 1.189 | 0.0835 | No | ||
| 21 | KCNA6 | 2779 | 1.189 | 0.0898 | No | ||
| 22 | CDC25A | 2883 | 1.156 | 0.0905 | No | ||
| 23 | SLC25A14 | 2907 | 1.150 | 0.0954 | No | ||
| 24 | ZCCHC8 | 2936 | 1.143 | 0.1000 | No | ||
| 25 | ARID4A | 3159 | 1.082 | 0.0938 | No | ||
| 26 | FANCC | 3282 | 1.049 | 0.0929 | No | ||
| 27 | MYC | 3332 | 1.038 | 0.0958 | No | ||
| 28 | STAG1 | 3363 | 1.029 | 0.0997 | No | ||
| 29 | FBXO5 | 3442 | 1.007 | 0.1009 | No | ||
| 30 | FANCG | 3565 | 0.972 | 0.0995 | No | ||
| 31 | PODN | 4234 | 0.816 | 0.0678 | No | ||
| 32 | KCNS2 | 4505 | 0.755 | 0.0572 | No | ||
| 33 | MRPL40 | 4525 | 0.751 | 0.0602 | No | ||
| 34 | RBL1 | 4596 | 0.738 | 0.0604 | No | ||
| 35 | CLSPN | 4710 | 0.715 | 0.0581 | No | ||
| 36 | MAP3K7 | 4950 | 0.669 | 0.0488 | No | ||
| 37 | ATF5 | 4990 | 0.661 | 0.0502 | No | ||
| 38 | EED | 5476 | 0.569 | 0.0270 | No | ||
| 39 | HIST1H2BK | 5549 | 0.555 | 0.0261 | No | ||
| 40 | USP52 | 5651 | 0.539 | 0.0235 | No | ||
| 41 | GRIA4 | 5773 | 0.514 | 0.0197 | No | ||
| 42 | YBX2 | 5849 | 0.498 | 0.0183 | No | ||
| 43 | OLFML3 | 6020 | 0.469 | 0.0117 | No | ||
| 44 | ZBTB4 | 6494 | 0.384 | -0.0119 | No | ||
| 45 | SFRS7 | 6610 | 0.368 | -0.0161 | No | ||
| 46 | GATA1 | 6753 | 0.347 | -0.0220 | No | ||
| 47 | H2AFV | 6966 | 0.310 | -0.0318 | No | ||
| 48 | HS6ST3 | 7114 | 0.280 | -0.0382 | No | ||
| 49 | BRMS1L | 7196 | 0.267 | -0.0412 | No | ||
| 50 | RQCD1 | 7217 | 0.263 | -0.0409 | No | ||
| 51 | HNRPD | 7296 | 0.251 | -0.0437 | No | ||
| 52 | ALDH6A1 | 7328 | 0.246 | -0.0441 | No | ||
| 53 | MCM3 | 7347 | 0.242 | -0.0438 | No | ||
| 54 | SOAT1 | 7360 | 0.240 | -0.0431 | No | ||
| 55 | ONECUT1 | 7392 | 0.235 | -0.0435 | No | ||
| 56 | STAG2 | 7498 | 0.220 | -0.0480 | No | ||
| 57 | STMN1 | 7625 | 0.200 | -0.0538 | No | ||
| 58 | PAPOLG | 7842 | 0.165 | -0.0646 | No | ||
| 59 | PEG3 | 8000 | 0.137 | -0.0724 | No | ||
| 60 | DNMT1 | 8419 | 0.077 | -0.0946 | No | ||
| 61 | SMAD6 | 8431 | 0.074 | -0.0948 | No | ||
| 62 | SEZ6 | 8434 | 0.073 | -0.0945 | No | ||
| 63 | RET | 8545 | 0.055 | -0.1002 | No | ||
| 64 | MCM8 | 8695 | 0.033 | -0.1081 | No | ||
| 65 | MDGA1 | 8826 | 0.012 | -0.1150 | No | ||
| 66 | NPR3 | 9043 | -0.023 | -0.1266 | No | ||
| 67 | PAQR4 | 9149 | -0.039 | -0.1321 | No | ||
| 68 | SLC9A7 | 9284 | -0.059 | -0.1390 | No | ||
| 69 | FMO4 | 9344 | -0.069 | -0.1419 | No | ||
| 70 | PRPS2 | 9380 | -0.075 | -0.1434 | No | ||
| 71 | H2AFZ | 9497 | -0.089 | -0.1492 | No | ||
| 72 | E2F7 | 9713 | -0.121 | -0.1602 | No | ||
| 73 | HNRPR | 10189 | -0.194 | -0.1849 | No | ||
| 74 | POLE2 | 10390 | -0.222 | -0.1945 | No | ||
| 75 | CASP8AP2 | 10666 | -0.271 | -0.2079 | No | ||
| 76 | PHC1 | 10676 | -0.273 | -0.2070 | No | ||
| 77 | E2F1 | 10726 | -0.280 | -0.2081 | No | ||
| 78 | ARHGAP6 | 10792 | -0.291 | -0.2101 | No | ||
| 79 | MSH5 | 11044 | -0.333 | -0.2219 | No | ||
| 80 | PAX6 | 11077 | -0.339 | -0.2218 | No | ||
| 81 | ATAD2 | 11078 | -0.339 | -0.2199 | No | ||
| 82 | SYNGR4 | 11108 | -0.344 | -0.2197 | No | ||
| 83 | RPS6KA5 | 11404 | -0.394 | -0.2335 | No | ||
| 84 | NRK | 11411 | -0.395 | -0.2317 | No | ||
| 85 | RPS20 | 11457 | -0.400 | -0.2320 | No | ||
| 86 | SEMA6A | 11585 | -0.421 | -0.2366 | No | ||
| 87 | PCSK1 | 11645 | -0.432 | -0.2375 | No | ||
| 88 | POLE4 | 11974 | -0.481 | -0.2527 | No | ||
| 89 | NUP62 | 12059 | -0.497 | -0.2545 | No | ||
| 90 | HIST1H4A | 12328 | -0.544 | -0.2661 | No | ||
| 91 | MCM2 | 12364 | -0.550 | -0.2651 | No | ||
| 92 | GPRC5B | 12414 | -0.558 | -0.2647 | No | ||
| 93 | GMNN | 12432 | -0.560 | -0.2626 | No | ||
| 94 | GABRB3 | 12521 | -0.579 | -0.2643 | No | ||
| 95 | NELL2 | 12555 | -0.587 | -0.2629 | No | ||
| 96 | SNRPD1 | 12627 | -0.601 | -0.2635 | No | ||
| 97 | MAPK6 | 12726 | -0.617 | -0.2655 | No | ||
| 98 | FHOD1 | 12789 | -0.626 | -0.2655 | No | ||
| 99 | PHF5A | 12862 | -0.639 | -0.2659 | No | ||
| 100 | SP3 | 12957 | -0.659 | -0.2675 | No | ||
| 101 | E2F3 | 13138 | -0.689 | -0.2735 | No | ||
| 102 | MCM7 | 13667 | -0.780 | -0.2979 | No | ||
| 103 | POLD3 | 13814 | -0.807 | -0.3015 | No | ||
| 104 | PPM1D | 13816 | -0.807 | -0.2972 | No | ||
| 105 | SYNCRIP | 13996 | -0.841 | -0.3024 | No | ||
| 106 | CDC6 | 14058 | -0.854 | -0.3011 | No | ||
| 107 | SUV39H1 | 14066 | -0.855 | -0.2968 | No | ||
| 108 | STK35 | 14143 | -0.871 | -0.2963 | No | ||
| 109 | ASXL2 | 14315 | -0.907 | -0.3007 | No | ||
| 110 | EIF4A1 | 14391 | -0.922 | -0.2998 | No | ||
| 111 | HIST1H2AH | 14632 | -0.975 | -0.3075 | No | ||
| 112 | POLR2A | 14928 | -1.043 | -0.3179 | No | ||
| 113 | NCL | 14983 | -1.054 | -0.3151 | No | ||
| 114 | EPHB1 | 15185 | -1.105 | -0.3201 | Yes | ||
| 115 | POU4F1 | 15224 | -1.118 | -0.3161 | Yes | ||
| 116 | PIM1 | 15228 | -1.119 | -0.3102 | Yes | ||
| 117 | NOLC1 | 15322 | -1.140 | -0.3091 | Yes | ||
| 118 | ACBD6 | 15338 | -1.146 | -0.3038 | Yes | ||
| 119 | ACO2 | 15376 | -1.155 | -0.2996 | Yes | ||
| 120 | HCN3 | 15580 | -1.209 | -0.3040 | Yes | ||
| 121 | AP4M1 | 15603 | -1.214 | -0.2987 | Yes | ||
| 122 | MAZ | 15802 | -1.268 | -0.3026 | Yes | ||
| 123 | HOXC10 | 15880 | -1.291 | -0.2998 | Yes | ||
| 124 | TOPBP1 | 15980 | -1.319 | -0.2981 | Yes | ||
| 125 | SUMO1 | 16060 | -1.345 | -0.2951 | Yes | ||
| 126 | AK2 | 16372 | -1.453 | -0.3041 | Yes | ||
| 127 | CDCA7 | 16385 | -1.459 | -0.2969 | Yes | ||
| 128 | PHF15 | 16475 | -1.495 | -0.2937 | Yes | ||
| 129 | GSPT1 | 16507 | -1.506 | -0.2873 | Yes | ||
| 130 | RRM2 | 16534 | -1.516 | -0.2805 | Yes | ||
| 131 | HTF9C | 16689 | -1.576 | -0.2804 | Yes | ||
| 132 | TYRO3 | 16693 | -1.579 | -0.2720 | Yes | ||
| 133 | PCNA | 16804 | -1.626 | -0.2692 | Yes | ||
| 134 | FKBP5 | 16880 | -1.666 | -0.2643 | Yes | ||
| 135 | UNG | 17016 | -1.732 | -0.2623 | Yes | ||
| 136 | HNRPUL1 | 17266 | -1.878 | -0.2657 | Yes | ||
| 137 | DCK | 17413 | -1.960 | -0.2630 | Yes | ||
| 138 | ILF3 | 17423 | -1.968 | -0.2529 | Yes | ||
| 139 | RANBP1 | 17543 | -2.058 | -0.2483 | Yes | ||
| 140 | LIG1 | 17601 | -2.111 | -0.2400 | Yes | ||
| 141 | SFRS10 | 17643 | -2.136 | -0.2307 | Yes | ||
| 142 | CORT | 17644 | -2.138 | -0.2192 | Yes | ||
| 143 | MSH2 | 17672 | -2.155 | -0.2091 | Yes | ||
| 144 | PRPS1 | 17743 | -2.219 | -0.2009 | Yes | ||
| 145 | USP37 | 17747 | -2.220 | -0.1891 | Yes | ||
| 146 | ING3 | 17779 | -2.247 | -0.1787 | Yes | ||
| 147 | AP1S1 | 17828 | -2.296 | -0.1689 | Yes | ||
| 148 | NASP | 17847 | -2.317 | -0.1574 | Yes | ||
| 149 | POLD1 | 17858 | -2.338 | -0.1454 | Yes | ||
| 150 | UGCGL1 | 18044 | -2.574 | -0.1415 | Yes | ||
| 151 | MCM4 | 18141 | -2.745 | -0.1319 | Yes | ||
| 152 | CBX3 | 18189 | -2.840 | -0.1192 | Yes | ||
| 153 | HNRPA2B1 | 18191 | -2.844 | -0.1039 | Yes | ||
| 154 | MCM6 | 18239 | -2.954 | -0.0906 | Yes | ||
| 155 | DMD | 18322 | -3.185 | -0.0778 | Yes | ||
| 156 | CDC5L | 18347 | -3.278 | -0.0615 | Yes | ||
| 157 | RBBP7 | 18365 | -3.348 | -0.0444 | Yes | ||
| 158 | EZH2 | 18393 | -3.451 | -0.0273 | Yes | ||
| 159 | MYH10 | 18433 | -3.640 | -0.0098 | Yes | ||
| 160 | TMPO | 18435 | -3.645 | 0.0098 | Yes |