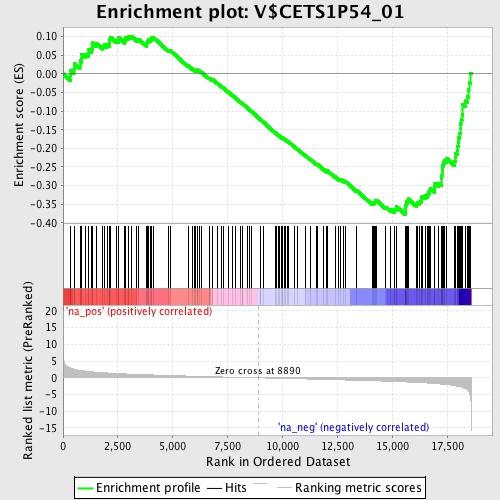
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$CETS1P54_01 |
| Enrichment Score (ES) | -0.3776615 |
| Normalized Enrichment Score (NES) | -1.6569344 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.5759126 |
| FWER p-Value | 0.678 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STX11 | 333 | 2.900 | -0.0035 | No | ||
| 2 | ARF3 | 350 | 2.862 | 0.0101 | No | ||
| 3 | RGS14 | 497 | 2.598 | 0.0152 | No | ||
| 4 | ARRB2 | 508 | 2.577 | 0.0277 | No | ||
| 5 | RRAS | 770 | 2.216 | 0.0247 | No | ||
| 6 | VPS52 | 774 | 2.214 | 0.0356 | No | ||
| 7 | SIPA1 | 847 | 2.148 | 0.0426 | No | ||
| 8 | STK10 | 852 | 2.142 | 0.0531 | No | ||
| 9 | RNF31 | 1034 | 1.986 | 0.0533 | No | ||
| 10 | EGFL7 | 1158 | 1.894 | 0.0562 | No | ||
| 11 | CHD2 | 1173 | 1.882 | 0.0649 | No | ||
| 12 | SPI1 | 1297 | 1.806 | 0.0673 | No | ||
| 13 | PIK3R4 | 1332 | 1.780 | 0.0744 | No | ||
| 14 | TNFSF13 | 1334 | 1.779 | 0.0833 | No | ||
| 15 | DR1 | 1502 | 1.676 | 0.0827 | No | ||
| 16 | NEDD8 | 1796 | 1.534 | 0.0745 | No | ||
| 17 | TEAD3 | 1872 | 1.503 | 0.0780 | No | ||
| 18 | AP1M1 | 2002 | 1.451 | 0.0783 | No | ||
| 19 | SEC24C | 2113 | 1.407 | 0.0795 | No | ||
| 20 | HMG20A | 2120 | 1.405 | 0.0862 | No | ||
| 21 | ACP2 | 2125 | 1.403 | 0.0930 | No | ||
| 22 | BAZ2A | 2148 | 1.397 | 0.0989 | No | ||
| 23 | NF1 | 2419 | 1.302 | 0.0908 | No | ||
| 24 | IPO7 | 2523 | 1.267 | 0.0916 | No | ||
| 25 | STK4 | 2540 | 1.261 | 0.0971 | No | ||
| 26 | CAST | 2816 | 1.177 | 0.0881 | No | ||
| 27 | CGGBP1 | 2834 | 1.172 | 0.0931 | No | ||
| 28 | HCST | 2857 | 1.165 | 0.0977 | No | ||
| 29 | DNAJA2 | 2959 | 1.137 | 0.0980 | No | ||
| 30 | TNF | 2999 | 1.123 | 0.1015 | No | ||
| 31 | ZDHHC5 | 3128 | 1.091 | 0.1001 | No | ||
| 32 | DNAJC1 | 3362 | 1.029 | 0.0926 | No | ||
| 33 | MUSK | 3453 | 1.005 | 0.0928 | No | ||
| 34 | RPS18 | 3790 | 0.918 | 0.0792 | No | ||
| 35 | BDKRB2 | 3816 | 0.913 | 0.0825 | No | ||
| 36 | RGL2 | 3829 | 0.910 | 0.0864 | No | ||
| 37 | TCF7 | 3887 | 0.897 | 0.0878 | No | ||
| 38 | GTF3C1 | 3894 | 0.895 | 0.0920 | No | ||
| 39 | TOMM22 | 4002 | 0.875 | 0.0906 | No | ||
| 40 | BCL2L1 | 4023 | 0.869 | 0.0939 | No | ||
| 41 | MEF2C | 4034 | 0.866 | 0.0977 | No | ||
| 42 | FBXO28 | 4108 | 0.849 | 0.0980 | No | ||
| 43 | B3GAT3 | 4824 | 0.691 | 0.0628 | No | ||
| 44 | DLST | 4880 | 0.681 | 0.0632 | No | ||
| 45 | GABRA1 | 5692 | 0.530 | 0.0219 | No | ||
| 46 | HTR2C | 5882 | 0.493 | 0.0142 | No | ||
| 47 | ATOH8 | 6005 | 0.471 | 0.0099 | No | ||
| 48 | LRP5 | 6043 | 0.465 | 0.0103 | No | ||
| 49 | EBNA1BP2 | 6116 | 0.453 | 0.0087 | No | ||
| 50 | ECGF1 | 6120 | 0.452 | 0.0108 | No | ||
| 51 | COX17 | 6212 | 0.435 | 0.0080 | No | ||
| 52 | CTCF | 6306 | 0.417 | 0.0051 | No | ||
| 53 | RAP2C | 6651 | 0.363 | -0.0117 | No | ||
| 54 | SFRS6 | 6689 | 0.356 | -0.0119 | No | ||
| 55 | GMPR2 | 6808 | 0.338 | -0.0166 | No | ||
| 56 | CHES1 | 6829 | 0.334 | -0.0160 | No | ||
| 57 | ZNF24 | 7020 | 0.300 | -0.0248 | No | ||
| 58 | EPN1 | 7197 | 0.267 | -0.0330 | No | ||
| 59 | HNRPD | 7296 | 0.251 | -0.0371 | No | ||
| 60 | SPRY2 | 7525 | 0.217 | -0.0483 | No | ||
| 61 | CD3E | 7728 | 0.184 | -0.0583 | No | ||
| 62 | ACTR2 | 7849 | 0.164 | -0.0640 | No | ||
| 63 | STARD13 | 8080 | 0.125 | -0.0758 | No | ||
| 64 | FMR1 | 8166 | 0.114 | -0.0799 | No | ||
| 65 | LYPLA2 | 8182 | 0.112 | -0.0801 | No | ||
| 66 | DNMT1 | 8419 | 0.077 | -0.0925 | No | ||
| 67 | PRKAG2 | 8515 | 0.059 | -0.0974 | No | ||
| 68 | RPLP2 | 8592 | 0.049 | -0.1012 | No | ||
| 69 | EXTL2 | 8986 | -0.014 | -0.1225 | No | ||
| 70 | TOMM70A | 8987 | -0.014 | -0.1224 | No | ||
| 71 | ZNF35 | 9131 | -0.036 | -0.1299 | No | ||
| 72 | PTPRN | 9670 | -0.115 | -0.1585 | No | ||
| 73 | XAB2 | 9742 | -0.125 | -0.1617 | No | ||
| 74 | MRPL52 | 9825 | -0.139 | -0.1655 | No | ||
| 75 | SHC3 | 9868 | -0.146 | -0.1670 | No | ||
| 76 | NRAS | 9973 | -0.163 | -0.1718 | No | ||
| 77 | PPP4C | 9991 | -0.165 | -0.1719 | No | ||
| 78 | OGG1 | 10014 | -0.170 | -0.1723 | No | ||
| 79 | CRADD | 10078 | -0.179 | -0.1748 | No | ||
| 80 | CSNK2B | 10120 | -0.184 | -0.1761 | No | ||
| 81 | MTIF2 | 10241 | -0.201 | -0.1816 | No | ||
| 82 | KRTCAP2 | 10283 | -0.208 | -0.1827 | No | ||
| 83 | TRIM44 | 10555 | -0.250 | -0.1962 | No | ||
| 84 | DPP8 | 10701 | -0.276 | -0.2026 | No | ||
| 85 | UQCRH | 11061 | -0.337 | -0.2204 | No | ||
| 86 | LIN28 | 11267 | -0.372 | -0.2296 | No | ||
| 87 | MAPKAPK2 | 11542 | -0.414 | -0.2424 | No | ||
| 88 | AKT1S1 | 11615 | -0.425 | -0.2441 | No | ||
| 89 | SYT5 | 11886 | -0.468 | -0.2564 | No | ||
| 90 | WDR8 | 12022 | -0.490 | -0.2613 | No | ||
| 91 | RPL26 | 12034 | -0.492 | -0.2594 | No | ||
| 92 | RNF12 | 12402 | -0.556 | -0.2765 | No | ||
| 93 | TBCC | 12573 | -0.590 | -0.2827 | No | ||
| 94 | MTPN | 12663 | -0.606 | -0.2845 | No | ||
| 95 | PSME2 | 12762 | -0.622 | -0.2867 | No | ||
| 96 | DDOST | 12848 | -0.637 | -0.2881 | No | ||
| 97 | PRKACB | 13382 | -0.726 | -0.3133 | No | ||
| 98 | FNTA | 14093 | -0.861 | -0.3474 | No | ||
| 99 | TRIM46 | 14145 | -0.872 | -0.3458 | No | ||
| 100 | HTATIP | 14210 | -0.884 | -0.3448 | No | ||
| 101 | LZIC | 14218 | -0.886 | -0.3407 | No | ||
| 102 | NRD1 | 14290 | -0.903 | -0.3401 | No | ||
| 103 | PSMD12 | 14706 | -0.990 | -0.3576 | No | ||
| 104 | U2AF2 | 14921 | -1.041 | -0.3639 | No | ||
| 105 | PAFAH1B2 | 15102 | -1.084 | -0.3682 | No | ||
| 106 | SHC1 | 15121 | -1.088 | -0.3637 | No | ||
| 107 | PPIL1 | 15190 | -1.106 | -0.3618 | No | ||
| 108 | GGN | 15191 | -1.107 | -0.3563 | No | ||
| 109 | MCTS1 | 15587 | -1.211 | -0.3716 | Yes | ||
| 110 | PSMC2 | 15595 | -1.212 | -0.3659 | Yes | ||
| 111 | NR1H2 | 15597 | -1.213 | -0.3598 | Yes | ||
| 112 | ITM2C | 15607 | -1.215 | -0.3542 | Yes | ||
| 113 | GNAI3 | 15629 | -1.219 | -0.3492 | Yes | ||
| 114 | FBXO3 | 15645 | -1.222 | -0.3439 | Yes | ||
| 115 | BAT4 | 15713 | -1.243 | -0.3412 | Yes | ||
| 116 | MTMR2 | 15730 | -1.247 | -0.3358 | Yes | ||
| 117 | GCNT3 | 16121 | -1.364 | -0.3501 | Yes | ||
| 118 | ZBTB11 | 16152 | -1.373 | -0.3448 | Yes | ||
| 119 | CSF3 | 16264 | -1.413 | -0.3437 | Yes | ||
| 120 | EN1 | 16316 | -1.435 | -0.3392 | Yes | ||
| 121 | RPL6 | 16318 | -1.435 | -0.3321 | Yes | ||
| 122 | FGFR4 | 16380 | -1.459 | -0.3280 | Yes | ||
| 123 | GSPT1 | 16507 | -1.506 | -0.3273 | Yes | ||
| 124 | PRKAG1 | 16600 | -1.541 | -0.3245 | Yes | ||
| 125 | CAP1 | 16637 | -1.558 | -0.3186 | Yes | ||
| 126 | BAG4 | 16686 | -1.574 | -0.3133 | Yes | ||
| 127 | STOML2 | 16722 | -1.592 | -0.3072 | Yes | ||
| 128 | ZNF22 | 16928 | -1.685 | -0.3098 | Yes | ||
| 129 | INSIG2 | 16937 | -1.690 | -0.3018 | Yes | ||
| 130 | BLNK | 16944 | -1.693 | -0.2936 | Yes | ||
| 131 | NOSIP | 17104 | -1.781 | -0.2932 | Yes | ||
| 132 | DDX47 | 17234 | -1.857 | -0.2909 | Yes | ||
| 133 | BAT3 | 17235 | -1.857 | -0.2815 | Yes | ||
| 134 | CALU | 17247 | -1.864 | -0.2727 | Yes | ||
| 135 | POLL | 17273 | -1.883 | -0.2646 | Yes | ||
| 136 | DMTF1 | 17277 | -1.886 | -0.2553 | Yes | ||
| 137 | FRAP1 | 17284 | -1.890 | -0.2461 | Yes | ||
| 138 | MKRN3 | 17319 | -1.908 | -0.2384 | Yes | ||
| 139 | MIR16 | 17372 | -1.936 | -0.2314 | Yes | ||
| 140 | HM13 | 17490 | -2.017 | -0.2276 | Yes | ||
| 141 | NASP | 17847 | -2.317 | -0.2353 | Yes | ||
| 142 | CUGBP1 | 17873 | -2.353 | -0.2248 | Yes | ||
| 143 | UBE1DC1 | 17885 | -2.369 | -0.2135 | Yes | ||
| 144 | SIRT3 | 17964 | -2.470 | -0.2053 | Yes | ||
| 145 | PRKACA | 17989 | -2.506 | -0.1939 | Yes | ||
| 146 | TRPC4AP | 18007 | -2.534 | -0.1821 | Yes | ||
| 147 | TBC1D17 | 18031 | -2.557 | -0.1705 | Yes | ||
| 148 | IL7R | 18074 | -2.641 | -0.1595 | Yes | ||
| 149 | AP1B1 | 18109 | -2.694 | -0.1478 | Yes | ||
| 150 | MRPL3 | 18113 | -2.704 | -0.1343 | Yes | ||
| 151 | LAG3 | 18136 | -2.736 | -0.1218 | Yes | ||
| 152 | SLC30A7 | 18182 | -2.833 | -0.1099 | Yes | ||
| 153 | HOXC4 | 18197 | -2.860 | -0.0963 | Yes | ||
| 154 | ARHGAP1 | 18198 | -2.863 | -0.0819 | Yes | ||
| 155 | SNRPB | 18325 | -3.196 | -0.0727 | Yes | ||
| 156 | PSMD13 | 18412 | -3.562 | -0.0594 | Yes | ||
| 157 | EIF5A | 18481 | -4.050 | -0.0427 | Yes | ||
| 158 | C19ORF20 | 18500 | -4.221 | -0.0224 | Yes | ||
| 159 | EPHA2 | 18572 | -5.699 | 0.0024 | Yes |