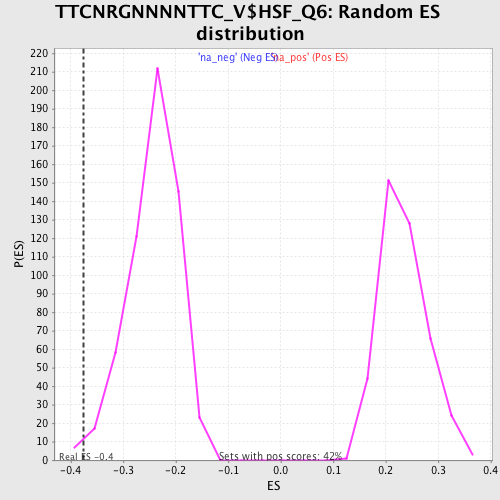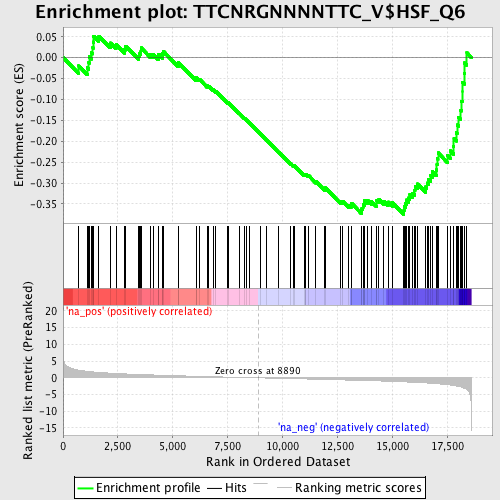
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTCNRGNNNNTTC_V$HSF_Q6 |
| Enrichment Score (ES) | -0.3752964 |
| Normalized Enrichment Score (NES) | -1.5411347 |
| Nominal p-value | 0.012006861 |
| FDR q-value | 0.34559152 |
| FWER p-Value | 0.977 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ECEL1 | 718 | 2.283 | -0.0197 | No | ||
| 2 | GPR4 | 1101 | 1.927 | -0.0242 | No | ||
| 3 | HSPA1L | 1171 | 1.886 | -0.0122 | No | ||
| 4 | SLC15A2 | 1187 | 1.875 | 0.0027 | No | ||
| 5 | C1RL | 1301 | 1.804 | 0.0117 | No | ||
| 6 | MAT2A | 1356 | 1.763 | 0.0235 | No | ||
| 7 | GATA6 | 1368 | 1.755 | 0.0376 | No | ||
| 8 | ATP2C1 | 1389 | 1.743 | 0.0511 | No | ||
| 9 | LHX1 | 1630 | 1.612 | 0.0516 | No | ||
| 10 | DAZL | 2151 | 1.397 | 0.0352 | No | ||
| 11 | TNFAIP8 | 2413 | 1.303 | 0.0320 | No | ||
| 12 | CDKL5 | 2806 | 1.181 | 0.0207 | No | ||
| 13 | ZNF148 | 2853 | 1.166 | 0.0279 | No | ||
| 14 | MUSK | 3453 | 1.005 | 0.0040 | No | ||
| 15 | XPO1 | 3496 | 0.991 | 0.0100 | No | ||
| 16 | UBE2B | 3545 | 0.976 | 0.0156 | No | ||
| 17 | UBB | 3549 | 0.975 | 0.0236 | No | ||
| 18 | PCTK2 | 3980 | 0.879 | 0.0077 | No | ||
| 19 | LRP8 | 4106 | 0.850 | 0.0080 | No | ||
| 20 | PVRL1 | 4339 | 0.791 | 0.0021 | No | ||
| 21 | UPF2 | 4343 | 0.790 | 0.0086 | No | ||
| 22 | FOXP1 | 4533 | 0.750 | 0.0046 | No | ||
| 23 | PMP22 | 4537 | 0.749 | 0.0107 | No | ||
| 24 | ZNF462 | 4586 | 0.740 | 0.0143 | No | ||
| 25 | CACNA1G | 5250 | 0.611 | -0.0164 | No | ||
| 26 | NR4A3 | 5261 | 0.609 | -0.0118 | No | ||
| 27 | TBPL1 | 6056 | 0.464 | -0.0509 | No | ||
| 28 | DNAJA1 | 6059 | 0.463 | -0.0471 | No | ||
| 29 | GJB2 | 6214 | 0.434 | -0.0518 | No | ||
| 30 | RAB39 | 6561 | 0.375 | -0.0673 | No | ||
| 31 | GPR50 | 6643 | 0.364 | -0.0687 | No | ||
| 32 | FOSB | 6847 | 0.331 | -0.0769 | No | ||
| 33 | MKNK2 | 6963 | 0.311 | -0.0805 | No | ||
| 34 | STAG2 | 7498 | 0.220 | -0.1075 | No | ||
| 35 | CANX | 7533 | 0.215 | -0.1075 | No | ||
| 36 | NEUROG1 | 8036 | 0.132 | -0.1335 | No | ||
| 37 | NR2E1 | 8273 | 0.099 | -0.1455 | No | ||
| 38 | TDGF1 | 8358 | 0.086 | -0.1493 | No | ||
| 39 | NFATC3 | 8491 | 0.063 | -0.1559 | No | ||
| 40 | HSPE1 | 8979 | -0.013 | -0.1821 | No | ||
| 41 | CEPT1 | 9277 | -0.058 | -0.1976 | No | ||
| 42 | MDS1 | 9832 | -0.140 | -0.2264 | No | ||
| 43 | YWHAE | 10376 | -0.220 | -0.2539 | No | ||
| 44 | EDN1 | 10501 | -0.241 | -0.2586 | No | ||
| 45 | NR2F1 | 10524 | -0.246 | -0.2577 | No | ||
| 46 | NODAL | 10980 | -0.322 | -0.2796 | No | ||
| 47 | LRRC2 | 11038 | -0.332 | -0.2799 | No | ||
| 48 | JAK1 | 11064 | -0.337 | -0.2784 | No | ||
| 49 | NUDT4 | 11190 | -0.358 | -0.2822 | No | ||
| 50 | CYP46A1 | 11523 | -0.411 | -0.2967 | No | ||
| 51 | CNTN6 | 11927 | -0.475 | -0.3145 | No | ||
| 52 | CRYGB | 11947 | -0.478 | -0.3115 | No | ||
| 53 | HOXA2 | 12634 | -0.603 | -0.3435 | No | ||
| 54 | YWHAG | 12725 | -0.617 | -0.3432 | No | ||
| 55 | DNAJB5 | 13009 | -0.670 | -0.3529 | No | ||
| 56 | STIP1 | 13126 | -0.687 | -0.3534 | No | ||
| 57 | E2F3 | 13138 | -0.689 | -0.3483 | No | ||
| 58 | SEMA6C | 13593 | -0.768 | -0.3664 | Yes | ||
| 59 | MAP6 | 13618 | -0.772 | -0.3612 | Yes | ||
| 60 | TWIST1 | 13668 | -0.780 | -0.3573 | Yes | ||
| 61 | CNGB3 | 13672 | -0.781 | -0.3510 | Yes | ||
| 62 | PAFAH1B1 | 13732 | -0.792 | -0.3475 | Yes | ||
| 63 | NNAT | 13747 | -0.795 | -0.3416 | Yes | ||
| 64 | CCT7 | 13884 | -0.820 | -0.3421 | Yes | ||
| 65 | PHF6 | 14052 | -0.853 | -0.3440 | Yes | ||
| 66 | CCT4 | 14295 | -0.903 | -0.3495 | Yes | ||
| 67 | CCT8 | 14296 | -0.904 | -0.3420 | Yes | ||
| 68 | EIF4A1 | 14391 | -0.922 | -0.3394 | Yes | ||
| 69 | HSPB2 | 14617 | -0.971 | -0.3434 | Yes | ||
| 70 | DMRTB1 | 14813 | -1.018 | -0.3454 | Yes | ||
| 71 | STARD8 | 15001 | -1.057 | -0.3467 | Yes | ||
| 72 | MYL1 | 15532 | -1.198 | -0.3653 | Yes | ||
| 73 | RBM6 | 15544 | -1.201 | -0.3558 | Yes | ||
| 74 | RANGAP1 | 15606 | -1.215 | -0.3490 | Yes | ||
| 75 | HSPA8 | 15640 | -1.221 | -0.3405 | Yes | ||
| 76 | RORA | 15746 | -1.250 | -0.3358 | Yes | ||
| 77 | NUTF2 | 15805 | -1.269 | -0.3283 | Yes | ||
| 78 | PTOV1 | 15928 | -1.305 | -0.3240 | Yes | ||
| 79 | RTN1 | 16028 | -1.334 | -0.3182 | Yes | ||
| 80 | PURA | 16040 | -1.338 | -0.3076 | Yes | ||
| 81 | PPID | 16154 | -1.374 | -0.3022 | Yes | ||
| 82 | EGR3 | 16523 | -1.514 | -0.3094 | Yes | ||
| 83 | TNXB | 16585 | -1.535 | -0.2998 | Yes | ||
| 84 | EIF4A2 | 16655 | -1.564 | -0.2905 | Yes | ||
| 85 | DPYSL2 | 16756 | -1.605 | -0.2825 | Yes | ||
| 86 | ABHD2 | 16827 | -1.641 | -0.2725 | Yes | ||
| 87 | AHSA1 | 17022 | -1.735 | -0.2685 | Yes | ||
| 88 | SERPINH1 | 17038 | -1.745 | -0.2547 | Yes | ||
| 89 | CHORDC1 | 17055 | -1.752 | -0.2409 | Yes | ||
| 90 | MORF4L2 | 17087 | -1.772 | -0.2278 | Yes | ||
| 91 | FGF10 | 17526 | -2.041 | -0.2344 | Yes | ||
| 92 | UBE2E3 | 17647 | -2.139 | -0.2230 | Yes | ||
| 93 | LMNB1 | 17798 | -2.267 | -0.2122 | Yes | ||
| 94 | INHBB | 17814 | -2.278 | -0.1939 | Yes | ||
| 95 | UBQLN1 | 17921 | -2.411 | -0.1795 | Yes | ||
| 96 | ACBD3 | 17959 | -2.460 | -0.1609 | Yes | ||
| 97 | HSPD1 | 18018 | -2.544 | -0.1428 | Yes | ||
| 98 | CRYAB | 18125 | -2.725 | -0.1257 | Yes | ||
| 99 | HSPH1 | 18163 | -2.779 | -0.1045 | Yes | ||
| 100 | CBX3 | 18189 | -2.840 | -0.0821 | Yes | ||
| 101 | HNRPA2B1 | 18191 | -2.844 | -0.0583 | Yes | ||
| 102 | ST13 | 18284 | -3.075 | -0.0376 | Yes | ||
| 103 | FKBP4 | 18304 | -3.127 | -0.0125 | Yes | ||
| 104 | MXD4 | 18405 | -3.503 | 0.0114 | Yes |