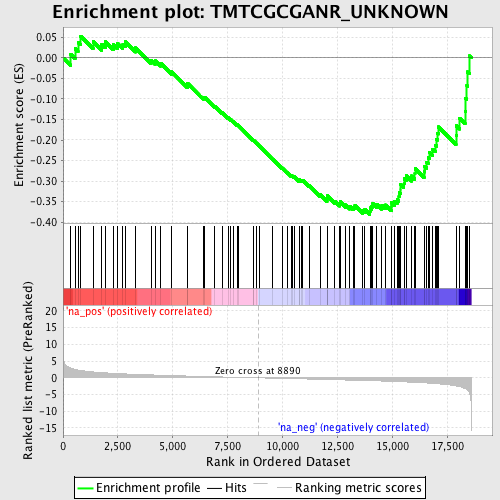
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TMTCGCGANR_UNKNOWN |
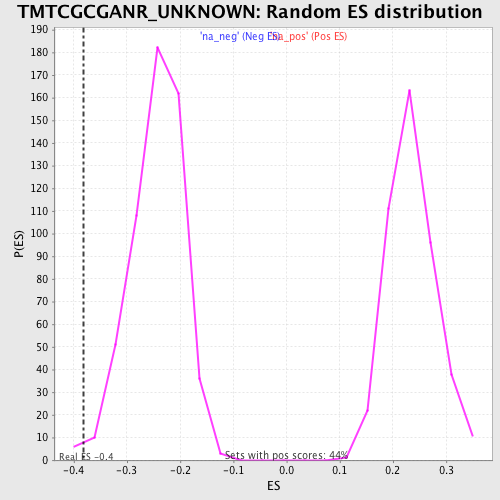
| Enrichment Score (ES) | -0.38162377 |
| Normalized Enrichment Score (NES) | -1.5636142 |
| Nominal p-value | 0.010752688 |
| FDR q-value | 0.5087895 |
| FWER p-Value | 0.956 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARF3 | 350 | 2.862 | 0.0090 | No | ||
| 2 | FGFR1OP2 | 546 | 2.526 | 0.0231 | No | ||
| 3 | ZNF346 | 682 | 2.330 | 0.0386 | No | ||
| 4 | MAPK14 | 804 | 2.181 | 0.0533 | No | ||
| 5 | ATF2 | 1367 | 1.756 | 0.0401 | No | ||
| 6 | DBR1 | 1762 | 1.547 | 0.0339 | No | ||
| 7 | PFKFB2 | 1926 | 1.480 | 0.0395 | No | ||
| 8 | LRSAM1 | 2294 | 1.337 | 0.0328 | No | ||
| 9 | SLC25A11 | 2474 | 1.283 | 0.0356 | No | ||
| 10 | ATF7 | 2725 | 1.203 | 0.0338 | No | ||
| 11 | PLOD2 | 2832 | 1.172 | 0.0396 | No | ||
| 12 | TTC14 | 3297 | 1.045 | 0.0247 | No | ||
| 13 | CHUK | 4007 | 0.874 | -0.0051 | No | ||
| 14 | TLK2 | 4193 | 0.829 | -0.0070 | No | ||
| 15 | PPP1R12A | 4432 | 0.774 | -0.0123 | No | ||
| 16 | MAP3K7 | 4950 | 0.669 | -0.0337 | No | ||
| 17 | KAZALD1 | 5660 | 0.536 | -0.0667 | No | ||
| 18 | LAMP1 | 5677 | 0.533 | -0.0624 | No | ||
| 19 | OFD1 | 6383 | 0.405 | -0.0965 | No | ||
| 20 | RPS15A | 6441 | 0.393 | -0.0957 | No | ||
| 21 | BCAS3 | 6913 | 0.319 | -0.1180 | No | ||
| 22 | DDX3X | 7250 | 0.258 | -0.1337 | No | ||
| 23 | SAP18 | 7517 | 0.218 | -0.1459 | No | ||
| 24 | WDR6 | 7631 | 0.200 | -0.1501 | No | ||
| 25 | RRAGA | 7744 | 0.182 | -0.1543 | No | ||
| 26 | VDAC3 | 7928 | 0.151 | -0.1627 | No | ||
| 27 | CCNJ | 7980 | 0.142 | -0.1641 | No | ||
| 28 | CD2AP | 8667 | 0.038 | -0.2008 | No | ||
| 29 | UBE2D3 | 8697 | 0.033 | -0.2020 | No | ||
| 30 | ANAPC4 | 8813 | 0.015 | -0.2081 | No | ||
| 31 | RPS7 | 8962 | -0.011 | -0.2160 | No | ||
| 32 | PIGN | 9544 | -0.096 | -0.2464 | No | ||
| 33 | PTPN4 | 9980 | -0.164 | -0.2683 | No | ||
| 34 | NDUFA11 | 10218 | -0.198 | -0.2791 | No | ||
| 35 | MRP63 | 10402 | -0.223 | -0.2868 | No | ||
| 36 | TROAP | 10461 | -0.233 | -0.2877 | No | ||
| 37 | CAMK2D | 10541 | -0.248 | -0.2895 | No | ||
| 38 | ATP5F1 | 10760 | -0.286 | -0.2985 | No | ||
| 39 | HSPA4 | 10780 | -0.290 | -0.2967 | No | ||
| 40 | COX11 | 10851 | -0.300 | -0.2976 | No | ||
| 41 | CNOT3 | 10908 | -0.310 | -0.2976 | No | ||
| 42 | MDH2 | 11216 | -0.363 | -0.3106 | No | ||
| 43 | RAB33B | 11715 | -0.441 | -0.3332 | No | ||
| 44 | SUPT5H | 12029 | -0.491 | -0.3453 | No | ||
| 45 | RPS6 | 12033 | -0.492 | -0.3407 | No | ||
| 46 | RPL26 | 12034 | -0.492 | -0.3359 | No | ||
| 47 | PSMB2 | 12385 | -0.553 | -0.3494 | No | ||
| 48 | NUDT2 | 12603 | -0.597 | -0.3553 | No | ||
| 49 | RPL17 | 12619 | -0.600 | -0.3502 | No | ||
| 50 | FUS | 12884 | -0.643 | -0.3582 | No | ||
| 51 | SLC12A2 | 13074 | -0.680 | -0.3618 | No | ||
| 52 | TLOC1 | 13239 | -0.704 | -0.3638 | No | ||
| 53 | ANKFY1 | 13300 | -0.713 | -0.3601 | No | ||
| 54 | NFYC | 13661 | -0.779 | -0.3719 | No | ||
| 55 | THAP7 | 13751 | -0.796 | -0.3689 | No | ||
| 56 | COX7B | 13987 | -0.839 | -0.3734 | Yes | ||
| 57 | GAS8 | 13993 | -0.841 | -0.3655 | Yes | ||
| 58 | SUV39H1 | 14066 | -0.855 | -0.3611 | Yes | ||
| 59 | EIF5 | 14116 | -0.866 | -0.3553 | Yes | ||
| 60 | CCT8 | 14296 | -0.904 | -0.3561 | Yes | ||
| 61 | LUC7L2 | 14529 | -0.952 | -0.3593 | Yes | ||
| 62 | SLC25A4 | 14691 | -0.987 | -0.3584 | Yes | ||
| 63 | MBTPS1 | 14960 | -1.048 | -0.3627 | Yes | ||
| 64 | THAP1 | 14964 | -1.049 | -0.3526 | Yes | ||
| 65 | DENR | 15106 | -1.084 | -0.3496 | Yes | ||
| 66 | POU4F1 | 15224 | -1.118 | -0.3450 | Yes | ||
| 67 | BZW1 | 15307 | -1.137 | -0.3384 | Yes | ||
| 68 | SMARCD2 | 15318 | -1.139 | -0.3278 | Yes | ||
| 69 | PRDX4 | 15380 | -1.157 | -0.3198 | Yes | ||
| 70 | SSR4 | 15381 | -1.157 | -0.3085 | Yes | ||
| 71 | ASH2L | 15536 | -1.199 | -0.3051 | Yes | ||
| 72 | UPF3B | 15545 | -1.201 | -0.2939 | Yes | ||
| 73 | RPL12 | 15639 | -1.220 | -0.2870 | Yes | ||
| 74 | ZZZ3 | 15885 | -1.292 | -0.2876 | Yes | ||
| 75 | TAS1R1 | 16036 | -1.337 | -0.2827 | Yes | ||
| 76 | PURA | 16040 | -1.338 | -0.2698 | Yes | ||
| 77 | STXBP4 | 16454 | -1.488 | -0.2776 | Yes | ||
| 78 | MATR3 | 16465 | -1.492 | -0.2636 | Yes | ||
| 79 | BANP | 16568 | -1.529 | -0.2542 | Yes | ||
| 80 | POLDIP3 | 16641 | -1.559 | -0.2428 | Yes | ||
| 81 | FSIP1 | 16697 | -1.582 | -0.2304 | Yes | ||
| 82 | RPL10A | 16840 | -1.646 | -0.2220 | Yes | ||
| 83 | RPS19 | 16973 | -1.705 | -0.2125 | Yes | ||
| 84 | SCAMP5 | 17024 | -1.736 | -0.1983 | Yes | ||
| 85 | PSMD5 | 17063 | -1.758 | -0.1832 | Yes | ||
| 86 | DGUOK | 17105 | -1.782 | -0.1680 | Yes | ||
| 87 | SEC63 | 17909 | -2.395 | -0.1880 | Yes | ||
| 88 | PRDX1 | 17924 | -2.412 | -0.1652 | Yes | ||
| 89 | IDH3G | 18076 | -2.642 | -0.1476 | Yes | ||
| 90 | DCTN2 | 18323 | -3.195 | -0.1297 | Yes | ||
| 91 | CDC5L | 18347 | -3.278 | -0.0990 | Yes | ||
| 92 | SFRS5 | 18402 | -3.494 | -0.0678 | Yes | ||
| 93 | HNRPK | 18432 | -3.635 | -0.0339 | Yes | ||
| 94 | UHRF2 | 18521 | -4.493 | 0.0051 | Yes |