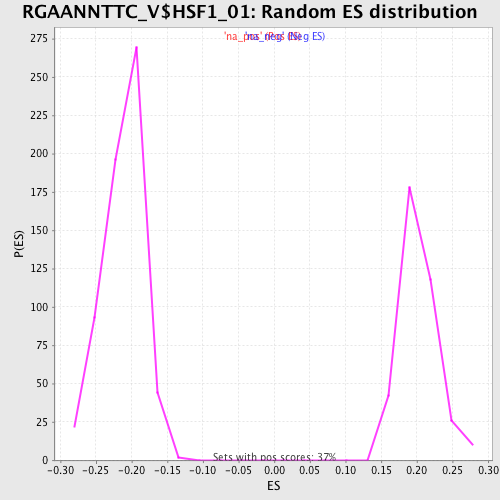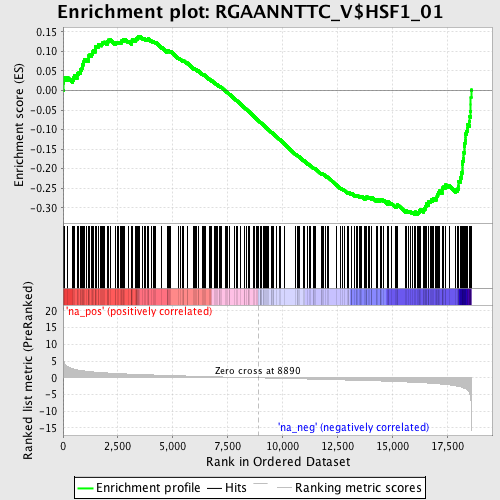
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RGAANNTTC_V$HSF1_01 |
| Enrichment Score (ES) | -0.3170405 |
| Normalized Enrichment Score (NES) | -1.50192 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.42352176 |
| FWER p-Value | 0.992 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | WNT10B | 11 | 7.646 | 0.0203 | No | ||
| 2 | IGFBP5 | 40 | 4.986 | 0.0325 | No | ||
| 3 | G0S2 | 189 | 3.431 | 0.0338 | No | ||
| 4 | RAB6B | 443 | 2.692 | 0.0274 | No | ||
| 5 | GCH1 | 484 | 2.616 | 0.0324 | No | ||
| 6 | ADAMTS2 | 511 | 2.571 | 0.0380 | No | ||
| 7 | CNTF | 637 | 2.405 | 0.0378 | No | ||
| 8 | CAMKK2 | 650 | 2.387 | 0.0436 | No | ||
| 9 | ECEL1 | 718 | 2.283 | 0.0462 | No | ||
| 10 | ADCY6 | 789 | 2.199 | 0.0484 | No | ||
| 11 | MAPK14 | 804 | 2.181 | 0.0537 | No | ||
| 12 | EDN2 | 855 | 2.140 | 0.0568 | No | ||
| 13 | BCL6 | 880 | 2.112 | 0.0613 | No | ||
| 14 | CYLD | 888 | 2.101 | 0.0666 | No | ||
| 15 | RASGRP2 | 920 | 2.073 | 0.0706 | No | ||
| 16 | IRF1 | 946 | 2.056 | 0.0749 | No | ||
| 17 | OGT | 952 | 2.052 | 0.0802 | No | ||
| 18 | IL6ST | 1083 | 1.941 | 0.0785 | No | ||
| 19 | GAD2 | 1154 | 1.897 | 0.0798 | No | ||
| 20 | HSPA1L | 1171 | 1.886 | 0.0841 | No | ||
| 21 | CHD2 | 1173 | 1.882 | 0.0892 | No | ||
| 22 | ABCD1 | 1221 | 1.848 | 0.0917 | No | ||
| 23 | EGF | 1302 | 1.804 | 0.0923 | No | ||
| 24 | FOXP2 | 1326 | 1.787 | 0.0960 | No | ||
| 25 | MAT2A | 1356 | 1.763 | 0.0992 | No | ||
| 26 | ATP2C1 | 1389 | 1.743 | 0.1022 | No | ||
| 27 | ENPP2 | 1464 | 1.696 | 0.1028 | No | ||
| 28 | DNAJB1 | 1468 | 1.694 | 0.1073 | No | ||
| 29 | LENEP | 1475 | 1.691 | 0.1116 | No | ||
| 30 | MOBP | 1526 | 1.662 | 0.1134 | No | ||
| 31 | SFRS1 | 1598 | 1.628 | 0.1140 | No | ||
| 32 | PSEN1 | 1622 | 1.618 | 0.1172 | No | ||
| 33 | DAPP1 | 1682 | 1.587 | 0.1183 | No | ||
| 34 | P4HA1 | 1769 | 1.544 | 0.1179 | No | ||
| 35 | OTX2 | 1779 | 1.542 | 0.1216 | No | ||
| 36 | ITGA8 | 1823 | 1.524 | 0.1234 | No | ||
| 37 | LRP1 | 1890 | 1.492 | 0.1239 | No | ||
| 38 | LUZP1 | 2009 | 1.449 | 0.1215 | No | ||
| 39 | ADAM15 | 2014 | 1.448 | 0.1252 | No | ||
| 40 | ADM | 2047 | 1.431 | 0.1274 | No | ||
| 41 | MBNL2 | 2088 | 1.415 | 0.1291 | No | ||
| 42 | DAZL | 2151 | 1.397 | 0.1295 | No | ||
| 43 | MESDC1 | 2392 | 1.309 | 0.1200 | No | ||
| 44 | SSBP3 | 2394 | 1.308 | 0.1235 | No | ||
| 45 | RALGDS | 2481 | 1.279 | 0.1223 | No | ||
| 46 | LHFPL2 | 2545 | 1.259 | 0.1224 | No | ||
| 47 | FHL1 | 2594 | 1.244 | 0.1231 | No | ||
| 48 | RFX3 | 2644 | 1.226 | 0.1238 | No | ||
| 49 | CNTFR | 2654 | 1.223 | 0.1267 | No | ||
| 50 | PIP5K1C | 2690 | 1.213 | 0.1281 | No | ||
| 51 | PLEKHB1 | 2757 | 1.193 | 0.1278 | No | ||
| 52 | BNC2 | 2759 | 1.193 | 0.1310 | No | ||
| 53 | OTP | 2813 | 1.179 | 0.1313 | No | ||
| 54 | TRERF1 | 2988 | 1.128 | 0.1249 | No | ||
| 55 | ZFX | 3115 | 1.093 | 0.1210 | No | ||
| 56 | TRPC6 | 3119 | 1.092 | 0.1239 | No | ||
| 57 | PNRC1 | 3133 | 1.089 | 0.1261 | No | ||
| 58 | HCFC1 | 3138 | 1.088 | 0.1289 | No | ||
| 59 | RPH3AL | 3168 | 1.080 | 0.1303 | No | ||
| 60 | FANCC | 3282 | 1.049 | 0.1270 | No | ||
| 61 | HIPK1 | 3322 | 1.040 | 0.1277 | No | ||
| 62 | PAX3 | 3325 | 1.039 | 0.1304 | No | ||
| 63 | GREB1 | 3340 | 1.035 | 0.1325 | No | ||
| 64 | PIP5K1A | 3378 | 1.025 | 0.1333 | No | ||
| 65 | BMP3 | 3406 | 1.017 | 0.1346 | No | ||
| 66 | MYOCD | 3421 | 1.014 | 0.1366 | No | ||
| 67 | DND1 | 3431 | 1.010 | 0.1389 | No | ||
| 68 | XPO1 | 3496 | 0.991 | 0.1381 | No | ||
| 69 | MAP4K4 | 3624 | 0.957 | 0.1338 | No | ||
| 70 | PRDM1 | 3691 | 0.941 | 0.1328 | No | ||
| 71 | SLC24A4 | 3774 | 0.921 | 0.1309 | No | ||
| 72 | ARL6IP2 | 3844 | 0.906 | 0.1296 | No | ||
| 73 | SPO11 | 3871 | 0.901 | 0.1306 | No | ||
| 74 | IL9 | 3880 | 0.899 | 0.1326 | No | ||
| 75 | MEF2C | 4034 | 0.866 | 0.1267 | No | ||
| 76 | COL6A3 | 4101 | 0.851 | 0.1254 | No | ||
| 77 | PTGIS | 4174 | 0.833 | 0.1238 | No | ||
| 78 | LAP3 | 4226 | 0.818 | 0.1232 | No | ||
| 79 | KCNS2 | 4505 | 0.755 | 0.1101 | No | ||
| 80 | COL4A6 | 4740 | 0.708 | 0.0993 | No | ||
| 81 | CHST2 | 4750 | 0.707 | 0.1007 | No | ||
| 82 | HIP1R | 4752 | 0.706 | 0.1026 | No | ||
| 83 | FSTL3 | 4783 | 0.700 | 0.1029 | No | ||
| 84 | NFATC4 | 4866 | 0.684 | 0.1003 | No | ||
| 85 | ATP6V1A | 4914 | 0.676 | 0.0996 | No | ||
| 86 | NR4A3 | 5261 | 0.609 | 0.0824 | No | ||
| 87 | TRDN | 5343 | 0.593 | 0.0796 | No | ||
| 88 | RORB | 5451 | 0.573 | 0.0753 | No | ||
| 89 | ASNS | 5480 | 0.568 | 0.0753 | No | ||
| 90 | EHD1 | 5507 | 0.563 | 0.0755 | No | ||
| 91 | JAM3 | 5652 | 0.539 | 0.0691 | No | ||
| 92 | CRISP1 | 5653 | 0.538 | 0.0705 | No | ||
| 93 | BCL9 | 5956 | 0.482 | 0.0554 | No | ||
| 94 | CCM2 | 5976 | 0.478 | 0.0557 | No | ||
| 95 | GRM1 | 6024 | 0.468 | 0.0544 | No | ||
| 96 | DNAJA1 | 6059 | 0.463 | 0.0538 | No | ||
| 97 | HES1 | 6151 | 0.447 | 0.0501 | No | ||
| 98 | PTPRA | 6153 | 0.447 | 0.0512 | No | ||
| 99 | PKN3 | 6355 | 0.409 | 0.0414 | No | ||
| 100 | BMPR2 | 6381 | 0.405 | 0.0411 | No | ||
| 101 | GPR17 | 6426 | 0.396 | 0.0398 | No | ||
| 102 | TYR | 6499 | 0.384 | 0.0369 | No | ||
| 103 | CREBBP | 6657 | 0.362 | 0.0294 | No | ||
| 104 | PERQ1 | 6700 | 0.356 | 0.0280 | No | ||
| 105 | STC1 | 6780 | 0.343 | 0.0247 | No | ||
| 106 | FLT4 | 6885 | 0.325 | 0.0199 | No | ||
| 107 | TRPS1 | 6947 | 0.313 | 0.0174 | No | ||
| 108 | EIF4ENIF1 | 6995 | 0.305 | 0.0157 | No | ||
| 109 | NEK6 | 7047 | 0.294 | 0.0137 | No | ||
| 110 | HS6ST3 | 7114 | 0.280 | 0.0109 | No | ||
| 111 | POLD4 | 7122 | 0.279 | 0.0113 | No | ||
| 112 | MRPS6 | 7183 | 0.269 | 0.0087 | No | ||
| 113 | GLRA3 | 7220 | 0.263 | 0.0075 | No | ||
| 114 | OSR2 | 7401 | 0.234 | -0.0017 | No | ||
| 115 | CHML | 7457 | 0.226 | -0.0041 | No | ||
| 116 | STAG2 | 7498 | 0.220 | -0.0057 | No | ||
| 117 | SFRS16 | 7583 | 0.207 | -0.0097 | No | ||
| 118 | CACNA1D | 7599 | 0.205 | -0.0099 | No | ||
| 119 | ISYNA1 | 7797 | 0.173 | -0.0202 | No | ||
| 120 | CLDN2 | 7908 | 0.155 | -0.0258 | No | ||
| 121 | B4GALT5 | 7956 | 0.146 | -0.0280 | No | ||
| 122 | STARD13 | 8080 | 0.125 | -0.0343 | No | ||
| 123 | ABHD3 | 8244 | 0.102 | -0.0429 | No | ||
| 124 | MAP2K1IP1 | 8263 | 0.101 | -0.0437 | No | ||
| 125 | PARVB | 8337 | 0.089 | -0.0474 | No | ||
| 126 | CXCR4 | 8439 | 0.072 | -0.0527 | No | ||
| 127 | IBRDC2 | 8441 | 0.072 | -0.0526 | No | ||
| 128 | NFATC3 | 8491 | 0.063 | -0.0551 | No | ||
| 129 | STX12 | 8699 | 0.033 | -0.0663 | No | ||
| 130 | BCAP31 | 8736 | 0.028 | -0.0682 | No | ||
| 131 | PLXNC1 | 8835 | 0.011 | -0.0735 | No | ||
| 132 | SP4 | 8859 | 0.006 | -0.0747 | No | ||
| 133 | SEMA3A | 8919 | -0.005 | -0.0779 | No | ||
| 134 | HSPE1 | 8979 | -0.013 | -0.0811 | No | ||
| 135 | SLC2A4 | 8997 | -0.015 | -0.0820 | No | ||
| 136 | SMYD1 | 9014 | -0.018 | -0.0828 | No | ||
| 137 | ADCY8 | 9027 | -0.021 | -0.0834 | No | ||
| 138 | HSPB1 | 9035 | -0.021 | -0.0837 | No | ||
| 139 | WDR12 | 9135 | -0.037 | -0.0890 | No | ||
| 140 | FBLN5 | 9139 | -0.038 | -0.0891 | No | ||
| 141 | PTHLH | 9180 | -0.043 | -0.0912 | No | ||
| 142 | FAP | 9210 | -0.047 | -0.0926 | No | ||
| 143 | CEPT1 | 9277 | -0.058 | -0.0961 | No | ||
| 144 | KNTC1 | 9328 | -0.066 | -0.0986 | No | ||
| 145 | DMRT1 | 9357 | -0.071 | -0.0999 | No | ||
| 146 | H2AFZ | 9497 | -0.089 | -0.1073 | No | ||
| 147 | TRIP10 | 9510 | -0.091 | -0.1077 | No | ||
| 148 | CACNA2D2 | 9538 | -0.095 | -0.1089 | No | ||
| 149 | SLC36A2 | 9607 | -0.105 | -0.1123 | No | ||
| 150 | MARK3 | 9608 | -0.105 | -0.1120 | No | ||
| 151 | MMP16 | 9716 | -0.122 | -0.1175 | No | ||
| 152 | SHC3 | 9868 | -0.146 | -0.1254 | No | ||
| 153 | EFNB3 | 9889 | -0.149 | -0.1261 | No | ||
| 154 | ADAMTS9 | 10107 | -0.181 | -0.1374 | No | ||
| 155 | COL4A5 | 10606 | -0.260 | -0.1639 | No | ||
| 156 | BDNF | 10661 | -0.271 | -0.1661 | No | ||
| 157 | CHST8 | 10663 | -0.271 | -0.1654 | No | ||
| 158 | PAK1IP1 | 10720 | -0.280 | -0.1677 | No | ||
| 159 | ARHGAP6 | 10792 | -0.291 | -0.1707 | No | ||
| 160 | HOXB8 | 10969 | -0.320 | -0.1795 | No | ||
| 161 | SCG2 | 10990 | -0.325 | -0.1797 | No | ||
| 162 | KCNQ1 | 11160 | -0.353 | -0.1879 | No | ||
| 163 | ANKRD22 | 11222 | -0.364 | -0.1903 | No | ||
| 164 | RIMS2 | 11271 | -0.373 | -0.1919 | No | ||
| 165 | AOC2 | 11423 | -0.396 | -0.1990 | No | ||
| 166 | DAPK1 | 11429 | -0.397 | -0.1982 | No | ||
| 167 | TREX1 | 11472 | -0.403 | -0.1994 | No | ||
| 168 | CYP46A1 | 11523 | -0.411 | -0.2010 | No | ||
| 169 | A2BP1 | 11787 | -0.453 | -0.2141 | No | ||
| 170 | CD9 | 11796 | -0.454 | -0.2133 | No | ||
| 171 | ZP3 | 11820 | -0.458 | -0.2133 | No | ||
| 172 | NR4A2 | 11857 | -0.463 | -0.2140 | No | ||
| 173 | PAK3 | 11980 | -0.482 | -0.2193 | No | ||
| 174 | PLXDC2 | 12037 | -0.492 | -0.2210 | No | ||
| 175 | HOXB5 | 12082 | -0.501 | -0.2221 | No | ||
| 176 | SIX4 | 12459 | -0.566 | -0.2410 | No | ||
| 177 | TLK1 | 12654 | -0.604 | -0.2499 | No | ||
| 178 | YWHAG | 12725 | -0.617 | -0.2521 | No | ||
| 179 | FGF9 | 12810 | -0.631 | -0.2549 | No | ||
| 180 | GAD1 | 12947 | -0.657 | -0.2606 | No | ||
| 181 | ARMCX3 | 13007 | -0.669 | -0.2619 | No | ||
| 182 | DNAJB5 | 13009 | -0.670 | -0.2602 | No | ||
| 183 | STIP1 | 13126 | -0.687 | -0.2646 | No | ||
| 184 | E2F3 | 13138 | -0.689 | -0.2633 | No | ||
| 185 | AMOT | 13153 | -0.691 | -0.2622 | No | ||
| 186 | ARHGAP21 | 13299 | -0.713 | -0.2681 | No | ||
| 187 | HCN4 | 13364 | -0.724 | -0.2697 | No | ||
| 188 | NRXN3 | 13391 | -0.728 | -0.2691 | No | ||
| 189 | ATP1B3 | 13418 | -0.732 | -0.2685 | No | ||
| 190 | HOXD13 | 13499 | -0.749 | -0.2708 | No | ||
| 191 | DBI | 13553 | -0.761 | -0.2716 | No | ||
| 192 | RORC | 13572 | -0.764 | -0.2705 | No | ||
| 193 | MAP6 | 13618 | -0.772 | -0.2708 | No | ||
| 194 | NNAT | 13747 | -0.795 | -0.2757 | No | ||
| 195 | RG9MTD2 | 13787 | -0.802 | -0.2756 | No | ||
| 196 | PPM1D | 13816 | -0.807 | -0.2749 | No | ||
| 197 | PPP2R5C | 13818 | -0.807 | -0.2727 | No | ||
| 198 | CASQ2 | 13846 | -0.812 | -0.2720 | No | ||
| 199 | NT5C3 | 13916 | -0.825 | -0.2735 | No | ||
| 200 | ALS2CR8 | 13958 | -0.833 | -0.2734 | No | ||
| 201 | FLNC | 14045 | -0.852 | -0.2758 | No | ||
| 202 | PHF6 | 14052 | -0.853 | -0.2738 | No | ||
| 203 | COL13A1 | 14070 | -0.856 | -0.2724 | No | ||
| 204 | CCT4 | 14295 | -0.903 | -0.2821 | No | ||
| 205 | CCT8 | 14296 | -0.904 | -0.2796 | No | ||
| 206 | GIPC2 | 14329 | -0.910 | -0.2789 | No | ||
| 207 | IL1F5 | 14443 | -0.932 | -0.2825 | No | ||
| 208 | PPT2 | 14446 | -0.933 | -0.2801 | No | ||
| 209 | NRF1 | 14479 | -0.940 | -0.2792 | No | ||
| 210 | CHD1 | 14507 | -0.946 | -0.2781 | No | ||
| 211 | HSPB2 | 14617 | -0.971 | -0.2814 | No | ||
| 212 | CREM | 14801 | -1.016 | -0.2886 | No | ||
| 213 | DMRTB1 | 14813 | -1.018 | -0.2864 | No | ||
| 214 | CUL2 | 14815 | -1.018 | -0.2837 | No | ||
| 215 | ELK3 | 14945 | -1.045 | -0.2879 | No | ||
| 216 | NOL4 | 15162 | -1.099 | -0.2966 | No | ||
| 217 | TPM2 | 15197 | -1.110 | -0.2954 | No | ||
| 218 | ATP1B2 | 15222 | -1.118 | -0.2937 | No | ||
| 219 | DMRTA1 | 15251 | -1.125 | -0.2921 | No | ||
| 220 | DMN | 15621 | -1.217 | -0.3089 | No | ||
| 221 | HSPA8 | 15640 | -1.221 | -0.3066 | No | ||
| 222 | FOXR1 | 15761 | -1.255 | -0.3097 | No | ||
| 223 | CACNA1A | 15815 | -1.272 | -0.3091 | No | ||
| 224 | GRM8 | 15917 | -1.302 | -0.3110 | No | ||
| 225 | RTN1 | 16028 | -1.334 | -0.3134 | Yes | ||
| 226 | PURA | 16040 | -1.338 | -0.3103 | Yes | ||
| 227 | PPID | 16154 | -1.374 | -0.3127 | Yes | ||
| 228 | FSIP2 | 16197 | -1.393 | -0.3112 | Yes | ||
| 229 | SRP54 | 16230 | -1.403 | -0.3091 | Yes | ||
| 230 | SLC35F3 | 16240 | -1.407 | -0.3057 | Yes | ||
| 231 | MAP3K7IP2 | 16296 | -1.427 | -0.3048 | Yes | ||
| 232 | MAGEH1 | 16412 | -1.469 | -0.3071 | Yes | ||
| 233 | SPAG9 | 16434 | -1.479 | -0.3042 | Yes | ||
| 234 | ELAVL4 | 16464 | -1.492 | -0.3017 | Yes | ||
| 235 | LHX3 | 16501 | -1.505 | -0.2995 | Yes | ||
| 236 | EGR3 | 16523 | -1.514 | -0.2965 | Yes | ||
| 237 | PLEC1 | 16547 | -1.521 | -0.2936 | Yes | ||
| 238 | PSMD10 | 16555 | -1.523 | -0.2898 | Yes | ||
| 239 | HOXB1 | 16649 | -1.561 | -0.2906 | Yes | ||
| 240 | EIF4A2 | 16655 | -1.564 | -0.2866 | Yes | ||
| 241 | EIF4G2 | 16666 | -1.569 | -0.2828 | Yes | ||
| 242 | GPR119 | 16746 | -1.602 | -0.2828 | Yes | ||
| 243 | ELF2 | 16767 | -1.611 | -0.2794 | Yes | ||
| 244 | ACTN2 | 16842 | -1.646 | -0.2790 | Yes | ||
| 245 | HIVEP2 | 16873 | -1.660 | -0.2761 | Yes | ||
| 246 | EOMES | 16961 | -1.700 | -0.2761 | Yes | ||
| 247 | DUSP1 | 17035 | -1.743 | -0.2754 | Yes | ||
| 248 | SERPINH1 | 17038 | -1.745 | -0.2707 | Yes | ||
| 249 | CHORDC1 | 17055 | -1.752 | -0.2668 | Yes | ||
| 250 | MORF4L2 | 17087 | -1.772 | -0.2636 | Yes | ||
| 251 | NOSIP | 17104 | -1.781 | -0.2596 | Yes | ||
| 252 | PACSIN3 | 17142 | -1.801 | -0.2567 | Yes | ||
| 253 | RBPMS | 17275 | -1.884 | -0.2587 | Yes | ||
| 254 | DMTF1 | 17277 | -1.886 | -0.2536 | Yes | ||
| 255 | GRIK1 | 17291 | -1.894 | -0.2491 | Yes | ||
| 256 | PIP5K3 | 17341 | -1.918 | -0.2465 | Yes | ||
| 257 | AAMP | 17411 | -1.958 | -0.2449 | Yes | ||
| 258 | CCT2 | 17445 | -1.990 | -0.2413 | Yes | ||
| 259 | CUTL1 | 17590 | -2.100 | -0.2434 | Yes | ||
| 260 | CYP1A1 | 17900 | -2.385 | -0.2537 | Yes | ||
| 261 | SMAD1 | 17991 | -2.512 | -0.2517 | Yes | ||
| 262 | PCDH10 | 18010 | -2.536 | -0.2458 | Yes | ||
| 263 | PDE6D | 18017 | -2.542 | -0.2391 | Yes | ||
| 264 | HSPD1 | 18018 | -2.544 | -0.2322 | Yes | ||
| 265 | FXYD2 | 18098 | -2.678 | -0.2291 | Yes | ||
| 266 | CRYAB | 18125 | -2.725 | -0.2231 | Yes | ||
| 267 | ABCC5 | 18147 | -2.750 | -0.2167 | Yes | ||
| 268 | HSPH1 | 18163 | -2.779 | -0.2099 | Yes | ||
| 269 | CBX3 | 18189 | -2.840 | -0.2035 | Yes | ||
| 270 | HNRPA2B1 | 18191 | -2.844 | -0.1958 | Yes | ||
| 271 | HSPB8 | 18192 | -2.845 | -0.1880 | Yes | ||
| 272 | RGS1 | 18213 | -2.890 | -0.1811 | Yes | ||
| 273 | MRPL45 | 18242 | -2.955 | -0.1746 | Yes | ||
| 274 | NTF5 | 18263 | -3.019 | -0.1674 | Yes | ||
| 275 | GSN | 18264 | -3.025 | -0.1591 | Yes | ||
| 276 | TNFAIP1 | 18283 | -3.074 | -0.1517 | Yes | ||
| 277 | ST13 | 18284 | -3.075 | -0.1432 | Yes | ||
| 278 | FKBP4 | 18304 | -3.127 | -0.1357 | Yes | ||
| 279 | DMD | 18322 | -3.185 | -0.1279 | Yes | ||
| 280 | JUN | 18337 | -3.236 | -0.1198 | Yes | ||
| 281 | GATA3 | 18340 | -3.255 | -0.1110 | Yes | ||
| 282 | CCT3 | 18392 | -3.448 | -0.1043 | Yes | ||
| 283 | ELOVL5 | 18418 | -3.580 | -0.0959 | Yes | ||
| 284 | MGLL | 18444 | -3.687 | -0.0872 | Yes | ||
| 285 | PLXNA2 | 18526 | -4.536 | -0.0791 | Yes | ||
| 286 | GNAS | 18538 | -4.797 | -0.0666 | Yes | ||
| 287 | EPHA2 | 18572 | -5.699 | -0.0528 | Yes | ||
| 288 | LMAN2L | 18588 | -6.510 | -0.0358 | Yes | ||
| 289 | CISH | 18589 | -6.631 | -0.0176 | Yes | ||
| 290 | SOCS2 | 18593 | -6.945 | 0.0013 | Yes |