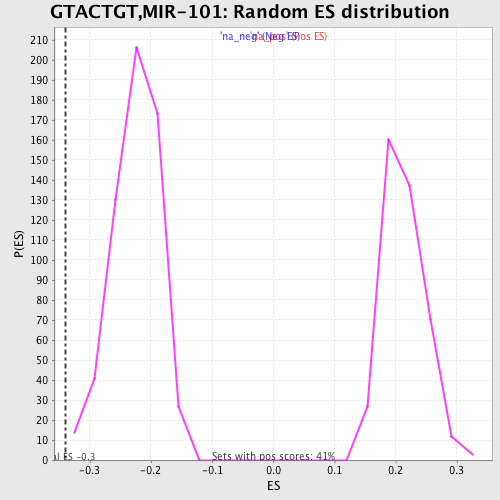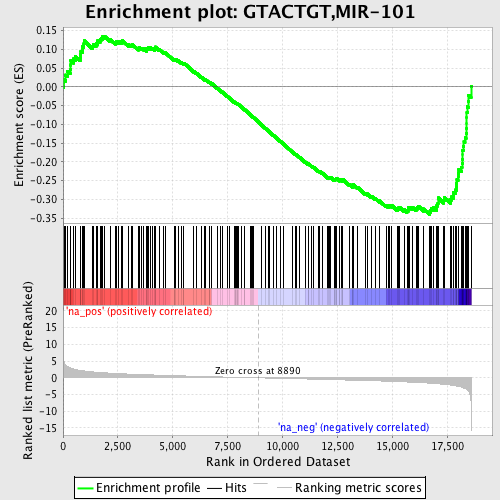
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GTACTGT,MIR-101 |
| Enrichment Score (ES) | -0.34013146 |
| Normalized Enrichment Score (NES) | -1.5173482 |
| Nominal p-value | 0.0016920473 |
| FDR q-value | 0.39749938 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IGFBP5 | 40 | 4.986 | 0.0189 | No | ||
| 2 | JDP2 | 114 | 3.916 | 0.0315 | No | ||
| 3 | PHLDA1 | 188 | 3.432 | 0.0420 | No | ||
| 4 | CDK5R1 | 320 | 2.949 | 0.0474 | No | ||
| 5 | WSB1 | 346 | 2.874 | 0.0581 | No | ||
| 6 | APP | 352 | 2.860 | 0.0700 | No | ||
| 7 | DNMT3A | 478 | 2.630 | 0.0743 | No | ||
| 8 | LASS2 | 552 | 2.514 | 0.0809 | No | ||
| 9 | CLCN3 | 778 | 2.210 | 0.0781 | No | ||
| 10 | ADCY6 | 789 | 2.199 | 0.0868 | No | ||
| 11 | MPPE1 | 810 | 2.175 | 0.0949 | No | ||
| 12 | PDE4A | 883 | 2.109 | 0.0999 | No | ||
| 13 | DDEF1 | 896 | 2.091 | 0.1081 | No | ||
| 14 | ARID1A | 947 | 2.056 | 0.1141 | No | ||
| 15 | OGT | 952 | 2.052 | 0.1225 | No | ||
| 16 | REV3L | 1346 | 1.770 | 0.1087 | No | ||
| 17 | HOXD4 | 1388 | 1.744 | 0.1138 | No | ||
| 18 | DR1 | 1502 | 1.676 | 0.1148 | No | ||
| 19 | PRPF4B | 1567 | 1.644 | 0.1182 | No | ||
| 20 | CDYL | 1585 | 1.636 | 0.1242 | No | ||
| 21 | SGPL1 | 1684 | 1.586 | 0.1256 | No | ||
| 22 | PURB | 1729 | 1.559 | 0.1298 | No | ||
| 23 | RRM1 | 1788 | 1.537 | 0.1332 | No | ||
| 24 | TEAD3 | 1872 | 1.503 | 0.1350 | No | ||
| 25 | BAZ2A | 2148 | 1.397 | 0.1260 | No | ||
| 26 | TGFBR1 | 2378 | 1.313 | 0.1191 | No | ||
| 27 | ERBB2IP | 2435 | 1.299 | 0.1216 | No | ||
| 28 | STK4 | 2540 | 1.261 | 0.1212 | No | ||
| 29 | PALM2 | 2661 | 1.221 | 0.1199 | No | ||
| 30 | PIP5K1C | 2690 | 1.213 | 0.1235 | No | ||
| 31 | PRKCE | 2978 | 1.130 | 0.1127 | No | ||
| 32 | SMARCD1 | 3098 | 1.097 | 0.1109 | No | ||
| 33 | MYST2 | 3156 | 1.083 | 0.1124 | No | ||
| 34 | PTGS2 | 3425 | 1.013 | 0.1021 | No | ||
| 35 | TAL1 | 3464 | 0.999 | 0.1043 | No | ||
| 36 | ANKRD17 | 3573 | 0.969 | 0.1025 | No | ||
| 37 | ZCCHC2 | 3660 | 0.948 | 0.1018 | No | ||
| 38 | PUM2 | 3800 | 0.917 | 0.0982 | No | ||
| 39 | RAP1B | 3802 | 0.917 | 0.1020 | No | ||
| 40 | USP47 | 3858 | 0.902 | 0.1028 | No | ||
| 41 | ATRX | 3903 | 0.894 | 0.1042 | No | ||
| 42 | SMARCA1 | 3973 | 0.880 | 0.1042 | No | ||
| 43 | TERF2 | 4070 | 0.857 | 0.1026 | No | ||
| 44 | NUDT11 | 4171 | 0.834 | 0.1007 | No | ||
| 45 | TLK2 | 4193 | 0.829 | 0.1031 | No | ||
| 46 | CEBPA | 4202 | 0.827 | 0.1061 | No | ||
| 47 | MYRIP | 4395 | 0.781 | 0.0990 | No | ||
| 48 | FBXW7 | 4571 | 0.743 | 0.0927 | No | ||
| 49 | ADAMTS10 | 4666 | 0.723 | 0.0906 | No | ||
| 50 | UBE2D2 | 5060 | 0.648 | 0.0720 | No | ||
| 51 | SOCS5 | 5078 | 0.644 | 0.0738 | No | ||
| 52 | AEBP2 | 5133 | 0.632 | 0.0736 | No | ||
| 53 | DLG5 | 5260 | 0.609 | 0.0693 | No | ||
| 54 | FBXW11 | 5399 | 0.583 | 0.0643 | No | ||
| 55 | EED | 5476 | 0.569 | 0.0626 | No | ||
| 56 | LRRC4 | 5506 | 0.563 | 0.0634 | No | ||
| 57 | BCL9 | 5956 | 0.482 | 0.0410 | No | ||
| 58 | SPRED1 | 6074 | 0.460 | 0.0366 | No | ||
| 59 | CTCF | 6306 | 0.417 | 0.0259 | No | ||
| 60 | DYRK1A | 6462 | 0.390 | 0.0191 | No | ||
| 61 | PACRG | 6509 | 0.382 | 0.0182 | No | ||
| 62 | RAP2C | 6651 | 0.363 | 0.0121 | No | ||
| 63 | RASD2 | 6677 | 0.358 | 0.0123 | No | ||
| 64 | STC1 | 6780 | 0.343 | 0.0082 | No | ||
| 65 | ZNF24 | 7020 | 0.300 | -0.0035 | No | ||
| 66 | UBE2A | 7195 | 0.267 | -0.0118 | No | ||
| 67 | DDX3X | 7250 | 0.258 | -0.0137 | No | ||
| 68 | STAG2 | 7498 | 0.220 | -0.0261 | No | ||
| 69 | PRKAA1 | 7579 | 0.207 | -0.0296 | No | ||
| 70 | ASPN | 7791 | 0.173 | -0.0403 | No | ||
| 71 | PAPOLG | 7842 | 0.165 | -0.0423 | No | ||
| 72 | ZFAND3 | 7876 | 0.161 | -0.0434 | No | ||
| 73 | CBL | 7889 | 0.160 | -0.0434 | No | ||
| 74 | RIN2 | 7968 | 0.144 | -0.0470 | No | ||
| 75 | PANK3 | 7970 | 0.144 | -0.0465 | No | ||
| 76 | CCNJ | 7980 | 0.142 | -0.0464 | No | ||
| 77 | DOT1L | 8132 | 0.119 | -0.0541 | No | ||
| 78 | DCBLD2 | 8276 | 0.099 | -0.0614 | No | ||
| 79 | MAK | 8287 | 0.097 | -0.0615 | No | ||
| 80 | HAS2 | 8527 | 0.058 | -0.0742 | No | ||
| 81 | ROBO2 | 8604 | 0.048 | -0.0782 | No | ||
| 82 | PPFIA4 | 8651 | 0.040 | -0.0805 | No | ||
| 83 | UBE2D3 | 8697 | 0.033 | -0.0828 | No | ||
| 84 | FZD4 | 9029 | -0.021 | -0.1007 | No | ||
| 85 | SOX11 | 9229 | -0.050 | -0.1112 | No | ||
| 86 | TULP4 | 9356 | -0.071 | -0.1178 | No | ||
| 87 | PLCG1 | 9400 | -0.078 | -0.1198 | No | ||
| 88 | MARK1 | 9592 | -0.104 | -0.1297 | No | ||
| 89 | GLRA2 | 9733 | -0.124 | -0.1368 | No | ||
| 90 | COL10A1 | 9895 | -0.150 | -0.1449 | No | ||
| 91 | EXOC5 | 10023 | -0.171 | -0.1510 | No | ||
| 92 | PURG | 10471 | -0.236 | -0.1743 | No | ||
| 93 | RAPH1 | 10600 | -0.259 | -0.1801 | No | ||
| 94 | CTTNBP2 | 10616 | -0.261 | -0.1799 | No | ||
| 95 | TMEFF1 | 10790 | -0.291 | -0.1880 | No | ||
| 96 | USP38 | 11043 | -0.332 | -0.2003 | No | ||
| 97 | JUNB | 11166 | -0.354 | -0.2054 | No | ||
| 98 | BICD2 | 11193 | -0.359 | -0.2053 | No | ||
| 99 | PCDH8 | 11333 | -0.382 | -0.2112 | No | ||
| 100 | NRK | 11411 | -0.395 | -0.2137 | No | ||
| 101 | GPR85 | 11649 | -0.432 | -0.2248 | No | ||
| 102 | MMP15 | 11695 | -0.439 | -0.2254 | No | ||
| 103 | ACCN2 | 11810 | -0.457 | -0.2296 | No | ||
| 104 | TXNDC4 | 12066 | -0.498 | -0.2413 | No | ||
| 105 | RAB1A | 12089 | -0.502 | -0.2404 | No | ||
| 106 | TRIB1 | 12157 | -0.514 | -0.2419 | No | ||
| 107 | PLXNB1 | 12194 | -0.522 | -0.2416 | No | ||
| 108 | MBNL1 | 12354 | -0.548 | -0.2479 | No | ||
| 109 | CDH11 | 12387 | -0.553 | -0.2473 | No | ||
| 110 | RIPK5 | 12407 | -0.557 | -0.2460 | No | ||
| 111 | THRB | 12449 | -0.564 | -0.2459 | No | ||
| 112 | SIX4 | 12459 | -0.566 | -0.2439 | No | ||
| 113 | DAG1 | 12593 | -0.595 | -0.2486 | No | ||
| 114 | SPOP | 12597 | -0.596 | -0.2463 | No | ||
| 115 | SOX9 | 12681 | -0.610 | -0.2482 | No | ||
| 116 | LRRN3 | 12746 | -0.620 | -0.2491 | No | ||
| 117 | KCNH7 | 12747 | -0.620 | -0.2465 | No | ||
| 118 | CDH5 | 13043 | -0.676 | -0.2596 | No | ||
| 119 | RAB5A | 13195 | -0.697 | -0.2649 | No | ||
| 120 | HNRPF | 13209 | -0.699 | -0.2626 | No | ||
| 121 | ZFHX4 | 13216 | -0.699 | -0.2600 | No | ||
| 122 | PPFIA1 | 13403 | -0.730 | -0.2670 | No | ||
| 123 | CPEB3 | 13778 | -0.800 | -0.2839 | No | ||
| 124 | QKI | 13866 | -0.815 | -0.2852 | No | ||
| 125 | FLRT3 | 14041 | -0.851 | -0.2910 | No | ||
| 126 | UBE2D1 | 14235 | -0.890 | -0.2977 | No | ||
| 127 | CPEB2 | 14415 | -0.926 | -0.3035 | No | ||
| 128 | LRRN1 | 14719 | -0.994 | -0.3158 | No | ||
| 129 | KIF1A | 14847 | -1.026 | -0.3183 | No | ||
| 130 | FBN2 | 14870 | -1.030 | -0.3152 | No | ||
| 131 | FGA | 14969 | -1.050 | -0.3160 | No | ||
| 132 | ETV5 | 15240 | -1.122 | -0.3259 | No | ||
| 133 | KPNB1 | 15267 | -1.128 | -0.3226 | No | ||
| 134 | POGZ | 15329 | -1.141 | -0.3211 | No | ||
| 135 | UBE2Q1 | 15537 | -1.199 | -0.3272 | No | ||
| 136 | ATXN1 | 15674 | -1.232 | -0.3294 | No | ||
| 137 | NEUROD1 | 15723 | -1.246 | -0.3268 | No | ||
| 138 | MTMR2 | 15730 | -1.247 | -0.3218 | No | ||
| 139 | SLC39A10 | 15804 | -1.269 | -0.3204 | No | ||
| 140 | CAV3 | 15938 | -1.309 | -0.3221 | No | ||
| 141 | NEGR1 | 16088 | -1.356 | -0.3245 | No | ||
| 142 | SLC1A1 | 16133 | -1.368 | -0.3211 | No | ||
| 143 | RANBP9 | 16191 | -1.390 | -0.3183 | No | ||
| 144 | DDX3Y | 16436 | -1.480 | -0.3253 | No | ||
| 145 | AP3S1 | 16711 | -1.588 | -0.3334 | Yes | ||
| 146 | CDK6 | 16749 | -1.603 | -0.3287 | Yes | ||
| 147 | NLK | 16771 | -1.613 | -0.3230 | Yes | ||
| 148 | FOS | 16885 | -1.668 | -0.3221 | Yes | ||
| 149 | NACA | 17027 | -1.737 | -0.3224 | Yes | ||
| 150 | DUSP1 | 17035 | -1.743 | -0.3154 | Yes | ||
| 151 | FLRT2 | 17081 | -1.768 | -0.3104 | Yes | ||
| 152 | NDFIP1 | 17091 | -1.773 | -0.3034 | Yes | ||
| 153 | RNF38 | 17092 | -1.776 | -0.2959 | Yes | ||
| 154 | EMP1 | 17350 | -1.922 | -0.3017 | Yes | ||
| 155 | GJA1 | 17371 | -1.935 | -0.2946 | Yes | ||
| 156 | BCL2L11 | 17655 | -2.143 | -0.3009 | Yes | ||
| 157 | ICK | 17685 | -2.165 | -0.2933 | Yes | ||
| 158 | ING3 | 17779 | -2.247 | -0.2889 | Yes | ||
| 159 | LMNB1 | 17798 | -2.267 | -0.2803 | Yes | ||
| 160 | GNB1 | 17881 | -2.364 | -0.2748 | Yes | ||
| 161 | FZD6 | 17916 | -2.402 | -0.2664 | Yes | ||
| 162 | PDE4D | 17934 | -2.425 | -0.2571 | Yes | ||
| 163 | SRPK2 | 17950 | -2.451 | -0.2476 | Yes | ||
| 164 | LRRFIP2 | 18001 | -2.528 | -0.2396 | Yes | ||
| 165 | BZW2 | 18011 | -2.539 | -0.2294 | Yes | ||
| 166 | FEM1C | 18036 | -2.564 | -0.2199 | Yes | ||
| 167 | ABCC5 | 18147 | -2.750 | -0.2142 | Yes | ||
| 168 | SLC30A7 | 18182 | -2.833 | -0.2041 | Yes | ||
| 169 | EYA1 | 18203 | -2.877 | -0.1930 | Yes | ||
| 170 | RGS1 | 18213 | -2.890 | -0.1813 | Yes | ||
| 171 | RXRB | 18215 | -2.892 | -0.1691 | Yes | ||
| 172 | CACNB2 | 18234 | -2.938 | -0.1577 | Yes | ||
| 173 | DDIT4 | 18270 | -3.044 | -0.1467 | Yes | ||
| 174 | TSC22D1 | 18318 | -3.170 | -0.1359 | Yes | ||
| 175 | RBBP7 | 18365 | -3.348 | -0.1243 | Yes | ||
| 176 | EZH2 | 18393 | -3.451 | -0.1111 | Yes | ||
| 177 | KHDRBS3 | 18400 | -3.487 | -0.0967 | Yes | ||
| 178 | SFRS5 | 18402 | -3.494 | -0.0820 | Yes | ||
| 179 | BTBD3 | 18406 | -3.513 | -0.0674 | Yes | ||
| 180 | MAGI1 | 18422 | -3.588 | -0.0530 | Yes | ||
| 181 | PREI3 | 18474 | -3.969 | -0.0390 | Yes | ||
| 182 | SLC19A2 | 18486 | -4.089 | -0.0223 | Yes | ||
| 183 | SOCS2 | 18593 | -6.945 | 0.0012 | Yes |