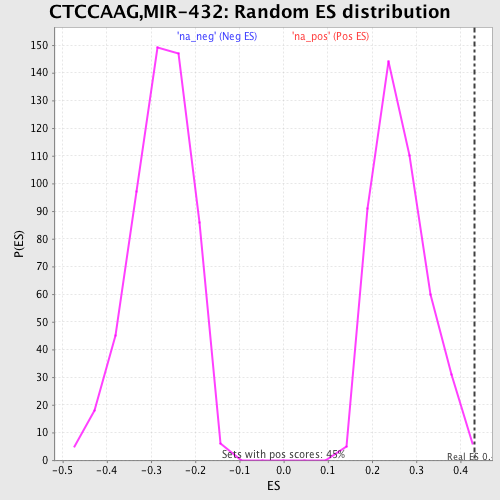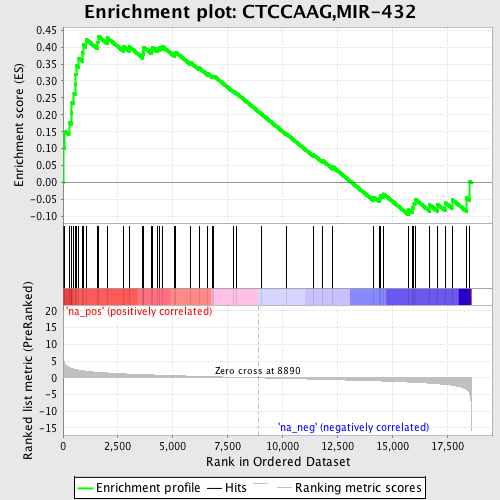
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CTCCAAG,MIR-432 |
| Enrichment Score (ES) | 0.43257016 |
| Normalized Enrichment Score (NES) | 1.6340408 |
| Nominal p-value | 0.008948546 |
| FDR q-value | 0.22671232 |
| FWER p-Value | 0.69 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHD3 | 4 | 8.830 | 0.1048 | Yes | ||
| 2 | EML4 | 76 | 4.265 | 0.1518 | Yes | ||
| 3 | RASGRP4 | 269 | 3.079 | 0.1780 | Yes | ||
| 4 | FN1 | 371 | 2.825 | 0.2062 | Yes | ||
| 5 | ESRRA | 402 | 2.765 | 0.2375 | Yes | ||
| 6 | PLAGL2 | 494 | 2.600 | 0.2635 | Yes | ||
| 7 | GRIA2 | 550 | 2.518 | 0.2905 | Yes | ||
| 8 | SDPR | 572 | 2.481 | 0.3189 | Yes | ||
| 9 | FURIN | 593 | 2.448 | 0.3470 | Yes | ||
| 10 | DAB2IP | 720 | 2.279 | 0.3673 | Yes | ||
| 11 | PDE4A | 883 | 2.109 | 0.3836 | Yes | ||
| 12 | NFAT5 | 907 | 2.083 | 0.4072 | Yes | ||
| 13 | BRPF1 | 1043 | 1.977 | 0.4234 | Yes | ||
| 14 | PRPF4B | 1567 | 1.644 | 0.4148 | Yes | ||
| 15 | SFRS1 | 1598 | 1.628 | 0.4326 | Yes | ||
| 16 | LUZP1 | 2009 | 1.449 | 0.4277 | No | ||
| 17 | CELSR2 | 2742 | 1.199 | 0.4026 | No | ||
| 18 | SPARC | 3013 | 1.119 | 0.4013 | No | ||
| 19 | AMFR | 3639 | 0.953 | 0.3790 | No | ||
| 20 | HSPA14 | 3664 | 0.947 | 0.3890 | No | ||
| 21 | ACE | 3676 | 0.945 | 0.3996 | No | ||
| 22 | CRB3 | 4033 | 0.866 | 0.3907 | No | ||
| 23 | ABCC3 | 4084 | 0.853 | 0.3982 | No | ||
| 24 | ABCA9 | 4304 | 0.796 | 0.3959 | No | ||
| 25 | CHKB | 4412 | 0.778 | 0.3994 | No | ||
| 26 | PMP22 | 4537 | 0.749 | 0.4016 | No | ||
| 27 | PDE1B | 5083 | 0.642 | 0.3799 | No | ||
| 28 | TLN1 | 5119 | 0.635 | 0.3855 | No | ||
| 29 | NR5A2 | 5803 | 0.508 | 0.3548 | No | ||
| 30 | RNF44 | 6199 | 0.437 | 0.3387 | No | ||
| 31 | CSE1L | 6595 | 0.370 | 0.3218 | No | ||
| 32 | ARL5B | 6816 | 0.337 | 0.3140 | No | ||
| 33 | PITPNC1 | 6876 | 0.326 | 0.3147 | No | ||
| 34 | HOXA5 | 7761 | 0.180 | 0.2692 | No | ||
| 35 | SMCR8 | 7905 | 0.155 | 0.2633 | No | ||
| 36 | EIF2C1 | 9019 | -0.019 | 0.2036 | No | ||
| 37 | CAPN3 | 10181 | -0.193 | 0.1433 | No | ||
| 38 | CPT1B | 11410 | -0.395 | 0.0819 | No | ||
| 39 | TRPC3 | 11829 | -0.459 | 0.0648 | No | ||
| 40 | HIF1AN | 12295 | -0.536 | 0.0462 | No | ||
| 41 | STK35 | 14143 | -0.871 | -0.0430 | No | ||
| 42 | CPEB2 | 14415 | -0.926 | -0.0465 | No | ||
| 43 | POLDIP2 | 14481 | -0.940 | -0.0389 | No | ||
| 44 | POLE3 | 14606 | -0.969 | -0.0340 | No | ||
| 45 | HMGA2 | 15731 | -1.248 | -0.0797 | No | ||
| 46 | FNTB | 15926 | -1.305 | -0.0746 | No | ||
| 47 | BACE1 | 15991 | -1.323 | -0.0623 | No | ||
| 48 | PURA | 16040 | -1.338 | -0.0490 | No | ||
| 49 | AP3S1 | 16711 | -1.588 | -0.0662 | No | ||
| 50 | SLMAP | 17079 | -1.767 | -0.0650 | No | ||
| 51 | HOMER2 | 17435 | -1.981 | -0.0605 | No | ||
| 52 | TRIM9 | 17727 | -2.201 | -0.0500 | No | ||
| 53 | DCTN3 | 18374 | -3.383 | -0.0445 | No | ||
| 54 | CA3 | 18542 | -4.835 | 0.0040 | No |