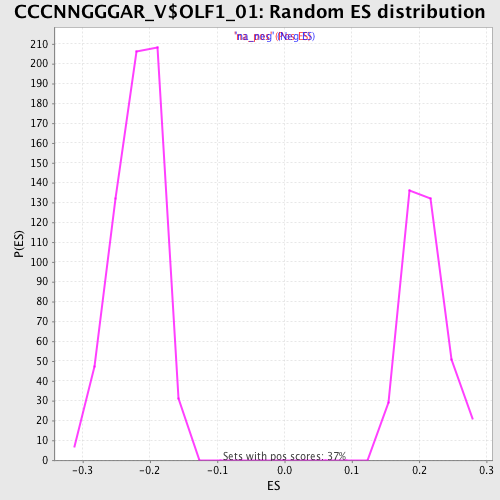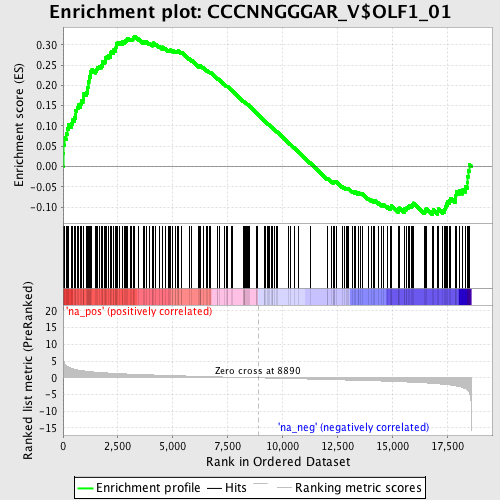
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CCCNNGGGAR_V$OLF1_01 |
| Enrichment Score (ES) | 0.32153448 |
| Normalized Enrichment Score (NES) | 1.540156 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2416003 |
| FWER p-Value | 0.927 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ROBO3 | 9 | 8.158 | 0.0328 | Yes | ||
| 2 | ABI3 | 33 | 5.394 | 0.0536 | Yes | ||
| 3 | AXL | 57 | 4.526 | 0.0708 | Yes | ||
| 4 | BCL9L | 141 | 3.722 | 0.0815 | Yes | ||
| 5 | MADCAM1 | 187 | 3.449 | 0.0931 | Yes | ||
| 6 | GCNT2 | 241 | 3.188 | 0.1032 | Yes | ||
| 7 | UBAP1 | 392 | 2.797 | 0.1065 | Yes | ||
| 8 | CLDN15 | 439 | 2.698 | 0.1150 | Yes | ||
| 9 | BCOR | 519 | 2.559 | 0.1212 | Yes | ||
| 10 | PLAG1 | 560 | 2.500 | 0.1292 | Yes | ||
| 11 | FCHSD2 | 568 | 2.488 | 0.1390 | Yes | ||
| 12 | MYOZ1 | 632 | 2.410 | 0.1454 | Yes | ||
| 13 | FLT1 | 685 | 2.327 | 0.1521 | Yes | ||
| 14 | ANXA9 | 809 | 2.175 | 0.1543 | Yes | ||
| 15 | ADCY4 | 824 | 2.162 | 0.1623 | Yes | ||
| 16 | TMEM24 | 909 | 2.078 | 0.1663 | Yes | ||
| 17 | HOXA11 | 940 | 2.059 | 0.1730 | Yes | ||
| 18 | ARID1A | 947 | 2.056 | 0.1811 | Yes | ||
| 19 | GNGT2 | 1050 | 1.969 | 0.1836 | Yes | ||
| 20 | TRIM8 | 1104 | 1.926 | 0.1886 | Yes | ||
| 21 | PRRX1 | 1117 | 1.920 | 0.1958 | Yes | ||
| 22 | TNIP1 | 1141 | 1.904 | 0.2023 | Yes | ||
| 23 | BAT2 | 1143 | 1.903 | 0.2100 | Yes | ||
| 24 | CD79B | 1181 | 1.878 | 0.2156 | Yes | ||
| 25 | THRA | 1188 | 1.875 | 0.2230 | Yes | ||
| 26 | HIC2 | 1256 | 1.831 | 0.2268 | Yes | ||
| 27 | H1F0 | 1266 | 1.824 | 0.2338 | Yes | ||
| 28 | APBA1 | 1314 | 1.799 | 0.2386 | Yes | ||
| 29 | ENPP2 | 1464 | 1.696 | 0.2374 | Yes | ||
| 30 | TNNT3 | 1537 | 1.657 | 0.2402 | Yes | ||
| 31 | SLC25A28 | 1570 | 1.643 | 0.2452 | Yes | ||
| 32 | GRIK3 | 1657 | 1.598 | 0.2471 | Yes | ||
| 33 | IL4I1 | 1735 | 1.557 | 0.2492 | Yes | ||
| 34 | TST | 1798 | 1.533 | 0.2521 | Yes | ||
| 35 | ADRB1 | 1802 | 1.532 | 0.2582 | Yes | ||
| 36 | LRP1 | 1890 | 1.492 | 0.2596 | Yes | ||
| 37 | FOXJ2 | 1932 | 1.477 | 0.2634 | Yes | ||
| 38 | COL18A1 | 1944 | 1.472 | 0.2688 | Yes | ||
| 39 | UBTF | 1999 | 1.452 | 0.2718 | Yes | ||
| 40 | ETV6 | 2086 | 1.415 | 0.2729 | Yes | ||
| 41 | BAZ2A | 2148 | 1.397 | 0.2753 | Yes | ||
| 42 | TBX21 | 2161 | 1.393 | 0.2803 | Yes | ||
| 43 | BMP4 | 2190 | 1.378 | 0.2844 | Yes | ||
| 44 | TNKS1BP1 | 2305 | 1.334 | 0.2837 | Yes | ||
| 45 | LPP | 2315 | 1.331 | 0.2886 | Yes | ||
| 46 | MESDC1 | 2392 | 1.309 | 0.2898 | Yes | ||
| 47 | GREM1 | 2408 | 1.304 | 0.2943 | Yes | ||
| 48 | HPCA | 2422 | 1.301 | 0.2989 | Yes | ||
| 49 | RGS3 | 2440 | 1.297 | 0.3033 | Yes | ||
| 50 | ABR | 2498 | 1.275 | 0.3054 | Yes | ||
| 51 | GNB2 | 2582 | 1.248 | 0.3060 | Yes | ||
| 52 | TIAM1 | 2696 | 1.211 | 0.3048 | Yes | ||
| 53 | ETV4 | 2723 | 1.203 | 0.3083 | Yes | ||
| 54 | COL23A1 | 2805 | 1.181 | 0.3087 | Yes | ||
| 55 | HCST | 2857 | 1.165 | 0.3107 | Yes | ||
| 56 | SLC25A14 | 2907 | 1.150 | 0.3127 | Yes | ||
| 57 | POU2AF1 | 2920 | 1.147 | 0.3168 | Yes | ||
| 58 | GJC1 | 3049 | 1.110 | 0.3143 | Yes | ||
| 59 | WDR13 | 3134 | 1.089 | 0.3142 | Yes | ||
| 60 | DUSP22 | 3185 | 1.076 | 0.3159 | Yes | ||
| 61 | TRAF4 | 3217 | 1.066 | 0.3186 | Yes | ||
| 62 | DAB1 | 3243 | 1.059 | 0.3215 | Yes | ||
| 63 | ELL2 | 3427 | 1.012 | 0.3157 | No | ||
| 64 | CHRDL1 | 3674 | 0.945 | 0.3062 | No | ||
| 65 | C1QL1 | 3689 | 0.942 | 0.3093 | No | ||
| 66 | IGF2R | 3787 | 0.919 | 0.3078 | No | ||
| 67 | NCDN | 3927 | 0.890 | 0.3039 | No | ||
| 68 | DPF1 | 4054 | 0.862 | 0.3005 | No | ||
| 69 | DUSP4 | 4089 | 0.852 | 0.3022 | No | ||
| 70 | LRP8 | 4106 | 0.850 | 0.3048 | No | ||
| 71 | RAB10 | 4224 | 0.818 | 0.3018 | No | ||
| 72 | CHCHD7 | 4394 | 0.781 | 0.2958 | No | ||
| 73 | UBE2N | 4513 | 0.753 | 0.2924 | No | ||
| 74 | DTX3 | 4527 | 0.750 | 0.2948 | No | ||
| 75 | VAT1 | 4653 | 0.725 | 0.2910 | No | ||
| 76 | FSTL3 | 4783 | 0.700 | 0.2868 | No | ||
| 77 | ATP2B3 | 4833 | 0.689 | 0.2870 | No | ||
| 78 | SNCB | 4874 | 0.683 | 0.2876 | No | ||
| 79 | IL11 | 4972 | 0.664 | 0.2850 | No | ||
| 80 | ATF5 | 4990 | 0.661 | 0.2868 | No | ||
| 81 | LRFN3 | 5099 | 0.638 | 0.2835 | No | ||
| 82 | LASP1 | 5138 | 0.631 | 0.2840 | No | ||
| 83 | LHX2 | 5197 | 0.620 | 0.2834 | No | ||
| 84 | JMJD1B | 5240 | 0.612 | 0.2836 | No | ||
| 85 | EPHB3 | 5253 | 0.610 | 0.2855 | No | ||
| 86 | NFKBIA | 5386 | 0.585 | 0.2807 | No | ||
| 87 | RHOQ | 5407 | 0.582 | 0.2820 | No | ||
| 88 | PSCD3 | 5746 | 0.518 | 0.2657 | No | ||
| 89 | PIAS1 | 5872 | 0.494 | 0.2610 | No | ||
| 90 | KCND1 | 6192 | 0.438 | 0.2454 | No | ||
| 91 | RNF44 | 6199 | 0.437 | 0.2469 | No | ||
| 92 | FBXL19 | 6243 | 0.429 | 0.2463 | No | ||
| 93 | DPYSL5 | 6255 | 0.428 | 0.2475 | No | ||
| 94 | REPIN1 | 6259 | 0.427 | 0.2490 | No | ||
| 95 | SMPD3 | 6384 | 0.404 | 0.2440 | No | ||
| 96 | COL11A2 | 6544 | 0.377 | 0.2369 | No | ||
| 97 | RAB39 | 6561 | 0.375 | 0.2375 | No | ||
| 98 | EPN3 | 6679 | 0.358 | 0.2326 | No | ||
| 99 | SLC41A1 | 6724 | 0.352 | 0.2317 | No | ||
| 100 | GRM7 | 7039 | 0.295 | 0.2158 | No | ||
| 101 | SDCCAG8 | 7049 | 0.294 | 0.2165 | No | ||
| 102 | GNAO1 | 7105 | 0.281 | 0.2147 | No | ||
| 103 | H3F3B | 7356 | 0.240 | 0.2021 | No | ||
| 104 | SCG3 | 7461 | 0.226 | 0.1974 | No | ||
| 105 | PTMA | 7484 | 0.222 | 0.1971 | No | ||
| 106 | STAG2 | 7498 | 0.220 | 0.1973 | No | ||
| 107 | ALOX12B | 7696 | 0.189 | 0.1873 | No | ||
| 108 | PCYT1B | 7718 | 0.185 | 0.1870 | No | ||
| 109 | VASP | 8198 | 0.109 | 0.1614 | No | ||
| 110 | PPP3CB | 8210 | 0.107 | 0.1612 | No | ||
| 111 | POGK | 8270 | 0.099 | 0.1584 | No | ||
| 112 | NR2E1 | 8273 | 0.099 | 0.1587 | No | ||
| 113 | PSME3 | 8288 | 0.097 | 0.1584 | No | ||
| 114 | NEO1 | 8298 | 0.096 | 0.1583 | No | ||
| 115 | SGK2 | 8357 | 0.086 | 0.1555 | No | ||
| 116 | CD72 | 8404 | 0.079 | 0.1533 | No | ||
| 117 | RHOG | 8430 | 0.075 | 0.1522 | No | ||
| 118 | MYOHD1 | 8473 | 0.067 | 0.1502 | No | ||
| 119 | LRRTM3 | 8822 | 0.013 | 0.1314 | No | ||
| 120 | MDGA1 | 8826 | 0.012 | 0.1313 | No | ||
| 121 | SP4 | 8859 | 0.006 | 0.1296 | No | ||
| 122 | STC2 | 9167 | -0.041 | 0.1130 | No | ||
| 123 | PTHLH | 9180 | -0.043 | 0.1126 | No | ||
| 124 | JARID2 | 9203 | -0.046 | 0.1116 | No | ||
| 125 | AKAP12 | 9294 | -0.061 | 0.1069 | No | ||
| 126 | SFRP2 | 9370 | -0.073 | 0.1032 | No | ||
| 127 | FKBP2 | 9383 | -0.075 | 0.1028 | No | ||
| 128 | MPST | 9488 | -0.088 | 0.0975 | No | ||
| 129 | ATP1A1 | 9532 | -0.094 | 0.0956 | No | ||
| 130 | ICAM1 | 9540 | -0.096 | 0.0956 | No | ||
| 131 | WNT7B | 9635 | -0.110 | 0.0909 | No | ||
| 132 | PIGW | 9744 | -0.126 | 0.0856 | No | ||
| 133 | JMJD2A | 9764 | -0.129 | 0.0851 | No | ||
| 134 | TRIM29 | 10256 | -0.204 | 0.0592 | No | ||
| 135 | PAX2 | 10364 | -0.219 | 0.0543 | No | ||
| 136 | CNTN2 | 10531 | -0.247 | 0.0463 | No | ||
| 137 | GPM6B | 10747 | -0.283 | 0.0358 | No | ||
| 138 | LIN28 | 11267 | -0.372 | 0.0091 | No | ||
| 139 | PLXDC2 | 12037 | -0.492 | -0.0306 | No | ||
| 140 | NUP62 | 12059 | -0.497 | -0.0298 | No | ||
| 141 | SLC30A5 | 12247 | -0.529 | -0.0378 | No | ||
| 142 | MTSS1 | 12310 | -0.540 | -0.0389 | No | ||
| 143 | CDON | 12312 | -0.541 | -0.0368 | No | ||
| 144 | SLC9A6 | 12355 | -0.548 | -0.0368 | No | ||
| 145 | LRRC20 | 12376 | -0.552 | -0.0356 | No | ||
| 146 | ADCYAP1 | 12438 | -0.561 | -0.0367 | No | ||
| 147 | NEUROD2 | 12716 | -0.616 | -0.0492 | No | ||
| 148 | HOXA6 | 12805 | -0.629 | -0.0514 | No | ||
| 149 | PDGFC | 12899 | -0.647 | -0.0538 | No | ||
| 150 | SP3 | 12957 | -0.659 | -0.0542 | No | ||
| 151 | ARMCX3 | 13007 | -0.669 | -0.0541 | No | ||
| 152 | MAG | 13208 | -0.698 | -0.0622 | No | ||
| 153 | ELAVL3 | 13298 | -0.712 | -0.0641 | No | ||
| 154 | EVI1 | 13312 | -0.715 | -0.0619 | No | ||
| 155 | TOM1L1 | 13448 | -0.738 | -0.0662 | No | ||
| 156 | EFS | 13450 | -0.738 | -0.0632 | No | ||
| 157 | RORC | 13572 | -0.764 | -0.0667 | No | ||
| 158 | DLL4 | 13636 | -0.775 | -0.0669 | No | ||
| 159 | EGR1 | 13899 | -0.822 | -0.0778 | No | ||
| 160 | TTYH1 | 14047 | -0.853 | -0.0823 | No | ||
| 161 | TRIB2 | 14161 | -0.874 | -0.0849 | No | ||
| 162 | MAGEL2 | 14185 | -0.879 | -0.0826 | No | ||
| 163 | FGF11 | 14382 | -0.920 | -0.0894 | No | ||
| 164 | LUC7L2 | 14529 | -0.952 | -0.0935 | No | ||
| 165 | MLLT6 | 14602 | -0.968 | -0.0935 | No | ||
| 166 | ZDHHC14 | 14774 | -1.008 | -0.0986 | No | ||
| 167 | DDR1 | 14915 | -1.040 | -0.1020 | No | ||
| 168 | ELK3 | 14945 | -1.045 | -0.0993 | No | ||
| 169 | CKM | 14977 | -1.053 | -0.0967 | No | ||
| 170 | RBM10 | 15268 | -1.128 | -0.1078 | No | ||
| 171 | CALM1 | 15269 | -1.128 | -0.1032 | No | ||
| 172 | ECM2 | 15333 | -1.143 | -0.1020 | No | ||
| 173 | UPF3B | 15545 | -1.201 | -0.1085 | No | ||
| 174 | TPM1 | 15553 | -1.203 | -0.1040 | No | ||
| 175 | BAI2 | 15630 | -1.219 | -0.1032 | No | ||
| 176 | THBS3 | 15671 | -1.230 | -0.1003 | No | ||
| 177 | HR | 15738 | -1.249 | -0.0988 | No | ||
| 178 | GRIN2D | 15788 | -1.264 | -0.0963 | No | ||
| 179 | HOXC10 | 15880 | -1.291 | -0.0960 | No | ||
| 180 | MAPKAP1 | 15915 | -1.301 | -0.0925 | No | ||
| 181 | LIMK2 | 15965 | -1.316 | -0.0898 | No | ||
| 182 | CHL1 | 16455 | -1.488 | -0.1103 | No | ||
| 183 | SSH2 | 16497 | -1.504 | -0.1064 | No | ||
| 184 | BANP | 16568 | -1.529 | -0.1039 | No | ||
| 185 | GRK5 | 16830 | -1.642 | -0.1114 | No | ||
| 186 | PACSIN1 | 16862 | -1.654 | -0.1063 | No | ||
| 187 | NF2 | 17064 | -1.758 | -0.1101 | No | ||
| 188 | NDFIP1 | 17091 | -1.773 | -0.1043 | No | ||
| 189 | GRIK1 | 17291 | -1.894 | -0.1073 | No | ||
| 190 | SYTL2 | 17396 | -1.947 | -0.1050 | No | ||
| 191 | AAMP | 17411 | -1.958 | -0.0978 | No | ||
| 192 | CHAT | 17454 | -1.995 | -0.0919 | No | ||
| 193 | PCBP4 | 17517 | -2.033 | -0.0870 | No | ||
| 194 | DLX1 | 17598 | -2.107 | -0.0827 | No | ||
| 195 | ICA1 | 17668 | -2.153 | -0.0777 | No | ||
| 196 | ADAMTS4 | 17870 | -2.350 | -0.0790 | No | ||
| 197 | MAGED2 | 17899 | -2.384 | -0.0708 | No | ||
| 198 | C1QTNF1 | 17908 | -2.394 | -0.0615 | No | ||
| 199 | PDAP1 | 18048 | -2.594 | -0.0584 | No | ||
| 200 | TENC1 | 18221 | -2.910 | -0.0559 | No | ||
| 201 | NDUFS2 | 18351 | -3.292 | -0.0495 | No | ||
| 202 | CA2 | 18421 | -3.585 | -0.0386 | No | ||
| 203 | SMARCE1 | 18436 | -3.658 | -0.0244 | No | ||
| 204 | MTX1 | 18464 | -3.877 | -0.0101 | No | ||
| 205 | HEBP2 | 18519 | -4.475 | 0.0053 | No |