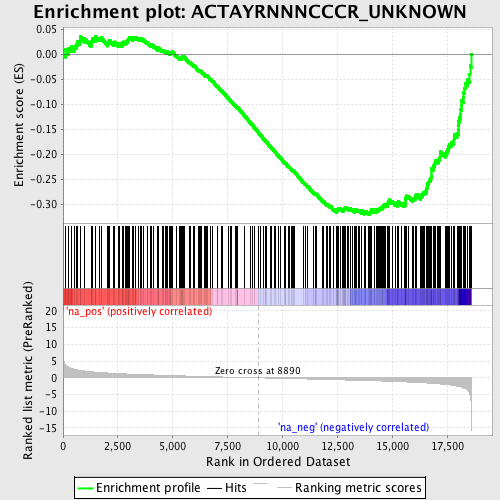
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ACTAYRNNNCCCR_UNKNOWN |
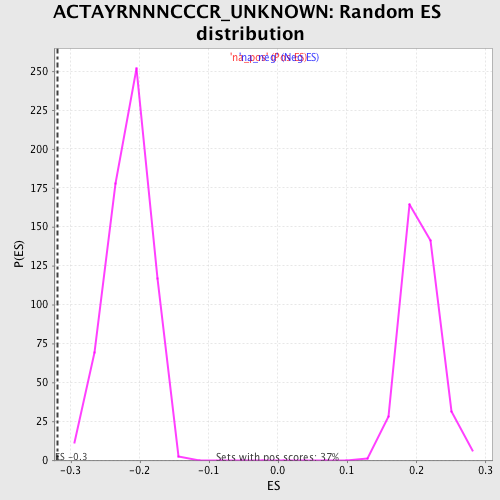
| Enrichment Score (ES) | -0.31979802 |
| Normalized Enrichment Score (NES) | -1.486564 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.4529103 |
| FWER p-Value | 0.996 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ORMDL3 | 94 | 4.068 | 0.0094 | No | ||
| 2 | COVA1 | 263 | 3.112 | 0.0113 | No | ||
| 3 | TBX6 | 373 | 2.818 | 0.0154 | No | ||
| 4 | NR1D1 | 531 | 2.540 | 0.0159 | No | ||
| 5 | TLE4 | 611 | 2.427 | 0.0203 | No | ||
| 6 | BIVM | 663 | 2.369 | 0.0259 | No | ||
| 7 | RRAS | 770 | 2.216 | 0.0281 | No | ||
| 8 | ZFYVE19 | 773 | 2.214 | 0.0359 | No | ||
| 9 | ITPR1 | 976 | 2.031 | 0.0321 | No | ||
| 10 | PHF12 | 1274 | 1.822 | 0.0224 | No | ||
| 11 | CDC45L | 1310 | 1.800 | 0.0269 | No | ||
| 12 | ZW10 | 1333 | 1.779 | 0.0321 | No | ||
| 13 | RAB8B | 1463 | 1.696 | 0.0311 | No | ||
| 14 | PDCD10 | 1470 | 1.693 | 0.0368 | No | ||
| 15 | DQX1 | 1647 | 1.601 | 0.0329 | No | ||
| 16 | RPIA | 1733 | 1.558 | 0.0338 | No | ||
| 17 | PPRC1 | 2043 | 1.433 | 0.0221 | No | ||
| 18 | TIAL1 | 2056 | 1.427 | 0.0266 | No | ||
| 19 | HMG20A | 2120 | 1.405 | 0.0281 | No | ||
| 20 | CYB561D2 | 2298 | 1.336 | 0.0233 | No | ||
| 21 | CDCA3 | 2352 | 1.319 | 0.0251 | No | ||
| 22 | IPO7 | 2523 | 1.267 | 0.0203 | No | ||
| 23 | TUSC4 | 2581 | 1.248 | 0.0217 | No | ||
| 24 | ZNF235 | 2687 | 1.214 | 0.0203 | No | ||
| 25 | NUDT13 | 2707 | 1.209 | 0.0236 | No | ||
| 26 | PLEKHB1 | 2757 | 1.193 | 0.0251 | No | ||
| 27 | HCFC1R1 | 2837 | 1.171 | 0.0250 | No | ||
| 28 | CDC25A | 2883 | 1.156 | 0.0267 | No | ||
| 29 | ZCCHC8 | 2936 | 1.143 | 0.0279 | No | ||
| 30 | MFN2 | 2963 | 1.135 | 0.0306 | No | ||
| 31 | PAFAH2 | 3002 | 1.123 | 0.0325 | No | ||
| 32 | BUB1B | 3033 | 1.114 | 0.0348 | No | ||
| 33 | GRWD1 | 3150 | 1.085 | 0.0324 | No | ||
| 34 | POP1 | 3190 | 1.074 | 0.0341 | No | ||
| 35 | QTRTD1 | 3281 | 1.049 | 0.0329 | No | ||
| 36 | ZCCHC7 | 3313 | 1.042 | 0.0349 | No | ||
| 37 | SUFU | 3445 | 1.006 | 0.0314 | No | ||
| 38 | CFL1 | 3509 | 0.989 | 0.0315 | No | ||
| 39 | FANCG | 3565 | 0.972 | 0.0320 | No | ||
| 40 | SFRS2 | 3669 | 0.946 | 0.0297 | No | ||
| 41 | GRK4 | 3828 | 0.910 | 0.0244 | No | ||
| 42 | FGFR1 | 3989 | 0.877 | 0.0188 | No | ||
| 43 | HSD17B4 | 4013 | 0.872 | 0.0206 | No | ||
| 44 | ITCH | 4124 | 0.846 | 0.0176 | No | ||
| 45 | SESN2 | 4295 | 0.799 | 0.0112 | No | ||
| 46 | PASK | 4325 | 0.793 | 0.0125 | No | ||
| 47 | NUP153 | 4336 | 0.792 | 0.0148 | No | ||
| 48 | UBE2N | 4513 | 0.753 | 0.0079 | No | ||
| 49 | GCN5L2 | 4553 | 0.747 | 0.0084 | No | ||
| 50 | TOP3A | 4649 | 0.725 | 0.0058 | No | ||
| 51 | STCH | 4732 | 0.710 | 0.0039 | No | ||
| 52 | SMARCAL1 | 4743 | 0.708 | 0.0059 | No | ||
| 53 | NUMA1 | 4841 | 0.688 | 0.0030 | No | ||
| 54 | MUS81 | 4881 | 0.681 | 0.0033 | No | ||
| 55 | PPP2R5B | 4897 | 0.679 | 0.0049 | No | ||
| 56 | MAX | 4952 | 0.669 | 0.0044 | No | ||
| 57 | SLC29A2 | 4964 | 0.666 | 0.0061 | No | ||
| 58 | INVS | 5161 | 0.627 | -0.0023 | No | ||
| 59 | SCAMP3 | 5302 | 0.601 | -0.0078 | No | ||
| 60 | MITF | 5336 | 0.594 | -0.0075 | No | ||
| 61 | TBN | 5379 | 0.586 | -0.0076 | No | ||
| 62 | FBXW11 | 5399 | 0.583 | -0.0066 | No | ||
| 63 | SERPINI1 | 5421 | 0.580 | -0.0057 | No | ||
| 64 | SMUG1 | 5424 | 0.579 | -0.0037 | No | ||
| 65 | SFXN5 | 5483 | 0.568 | -0.0049 | No | ||
| 66 | DFFA | 5509 | 0.563 | -0.0042 | No | ||
| 67 | DTX2 | 5742 | 0.519 | -0.0150 | No | ||
| 68 | RPP21 | 5783 | 0.512 | -0.0154 | No | ||
| 69 | RDH12 | 5920 | 0.487 | -0.0210 | No | ||
| 70 | KIAA0427 | 5965 | 0.481 | -0.0217 | No | ||
| 71 | RAB3C | 6175 | 0.441 | -0.0315 | No | ||
| 72 | DDX25 | 6236 | 0.430 | -0.0333 | No | ||
| 73 | DPYSL5 | 6255 | 0.428 | -0.0327 | No | ||
| 74 | UFD1L | 6299 | 0.418 | -0.0336 | No | ||
| 75 | NCOA3 | 6449 | 0.392 | -0.0403 | No | ||
| 76 | PACRG | 6509 | 0.382 | -0.0421 | No | ||
| 77 | PHF13 | 6554 | 0.376 | -0.0432 | No | ||
| 78 | WDFY2 | 6573 | 0.374 | -0.0428 | No | ||
| 79 | KATNAL1 | 6723 | 0.353 | -0.0497 | No | ||
| 80 | VKORC1 | 6812 | 0.337 | -0.0533 | No | ||
| 81 | ZNF24 | 7020 | 0.300 | -0.0635 | No | ||
| 82 | SDCCAG8 | 7049 | 0.294 | -0.0640 | No | ||
| 83 | BRMS1L | 7196 | 0.267 | -0.0710 | No | ||
| 84 | IRF2BP1 | 7272 | 0.254 | -0.0741 | No | ||
| 85 | ATP6V1F | 7515 | 0.218 | -0.0865 | No | ||
| 86 | STMN1 | 7625 | 0.200 | -0.0918 | No | ||
| 87 | USP48 | 7687 | 0.190 | -0.0944 | No | ||
| 88 | IPO13 | 7836 | 0.167 | -0.1019 | No | ||
| 89 | SMCR8 | 7905 | 0.155 | -0.1050 | No | ||
| 90 | FEN1 | 7949 | 0.147 | -0.1068 | No | ||
| 91 | SIRT2 | 7954 | 0.147 | -0.1065 | No | ||
| 92 | HSPC171 | 7966 | 0.145 | -0.1066 | No | ||
| 93 | PANK3 | 7970 | 0.144 | -0.1063 | No | ||
| 94 | UBL3 | 8267 | 0.100 | -0.1220 | No | ||
| 95 | BUB3 | 8562 | 0.053 | -0.1379 | No | ||
| 96 | RPS11 | 8627 | 0.044 | -0.1412 | No | ||
| 97 | WRN | 8705 | 0.032 | -0.1453 | No | ||
| 98 | CTF1 | 8892 | -0.000 | -0.1554 | No | ||
| 99 | PPP1R7 | 9007 | -0.017 | -0.1616 | No | ||
| 100 | NFKBIB | 9110 | -0.033 | -0.1670 | No | ||
| 101 | JARID2 | 9203 | -0.046 | -0.1718 | No | ||
| 102 | COX7C | 9208 | -0.047 | -0.1719 | No | ||
| 103 | SNX16 | 9275 | -0.058 | -0.1753 | No | ||
| 104 | USP5 | 9438 | -0.082 | -0.1838 | No | ||
| 105 | TRIP10 | 9510 | -0.091 | -0.1874 | No | ||
| 106 | MAPBPIP | 9642 | -0.111 | -0.1941 | No | ||
| 107 | POLR1B | 9661 | -0.113 | -0.1947 | No | ||
| 108 | BTRC | 9809 | -0.136 | -0.2022 | No | ||
| 109 | BCKDK | 9918 | -0.155 | -0.2075 | No | ||
| 110 | RPL15 | 10109 | -0.182 | -0.2172 | No | ||
| 111 | SNX1 | 10117 | -0.183 | -0.2169 | No | ||
| 112 | KCNE4 | 10128 | -0.185 | -0.2168 | No | ||
| 113 | SERTAD3 | 10249 | -0.203 | -0.2226 | No | ||
| 114 | RAD51L1 | 10322 | -0.213 | -0.2258 | No | ||
| 115 | RPS6KB2 | 10407 | -0.223 | -0.2296 | No | ||
| 116 | PURG | 10471 | -0.236 | -0.2322 | No | ||
| 117 | NKIRAS1 | 10510 | -0.244 | -0.2334 | No | ||
| 118 | WDTC1 | 10567 | -0.251 | -0.2355 | No | ||
| 119 | RAB5C | 10946 | -0.316 | -0.2550 | No | ||
| 120 | SYT3 | 11041 | -0.332 | -0.2589 | No | ||
| 121 | BRCA1 | 11152 | -0.351 | -0.2637 | No | ||
| 122 | HACE1 | 11407 | -0.394 | -0.2761 | No | ||
| 123 | ITSN1 | 11494 | -0.407 | -0.2793 | No | ||
| 124 | PA2G4 | 11499 | -0.407 | -0.2781 | No | ||
| 125 | APBA3 | 11549 | -0.415 | -0.2793 | No | ||
| 126 | NTAN1 | 11821 | -0.458 | -0.2924 | No | ||
| 127 | TTLL4 | 11888 | -0.468 | -0.2943 | No | ||
| 128 | PRPF4 | 12017 | -0.489 | -0.2996 | No | ||
| 129 | RABL3 | 12056 | -0.496 | -0.2999 | No | ||
| 130 | CBLL1 | 12151 | -0.512 | -0.3032 | No | ||
| 131 | ZNF8 | 12179 | -0.519 | -0.3028 | No | ||
| 132 | RNF10 | 12339 | -0.546 | -0.3095 | No | ||
| 133 | RPP40 | 12465 | -0.568 | -0.3143 | No | ||
| 134 | RDH11 | 12481 | -0.573 | -0.3131 | No | ||
| 135 | CDC26 | 12487 | -0.574 | -0.3113 | No | ||
| 136 | DPM3 | 12498 | -0.575 | -0.3098 | No | ||
| 137 | CALR3 | 12522 | -0.579 | -0.3090 | No | ||
| 138 | GDAP1 | 12536 | -0.582 | -0.3076 | No | ||
| 139 | GSK3A | 12568 | -0.589 | -0.3072 | No | ||
| 140 | SHMT2 | 12628 | -0.602 | -0.3083 | No | ||
| 141 | YWHAG | 12725 | -0.617 | -0.3113 | No | ||
| 142 | TRIM23 | 12758 | -0.621 | -0.3108 | No | ||
| 143 | CYP39A1 | 12793 | -0.627 | -0.3104 | No | ||
| 144 | FDPS | 12803 | -0.629 | -0.3087 | No | ||
| 145 | MAP3K10 | 12811 | -0.631 | -0.3068 | No | ||
| 146 | MAP2K6 | 12872 | -0.641 | -0.3078 | No | ||
| 147 | CDK9 | 12891 | -0.645 | -0.3065 | No | ||
| 148 | DHX30 | 12942 | -0.655 | -0.3069 | No | ||
| 149 | PTDSS2 | 13021 | -0.671 | -0.3087 | No | ||
| 150 | SLC25A27 | 13091 | -0.682 | -0.3101 | No | ||
| 151 | PSMD11 | 13094 | -0.682 | -0.3077 | No | ||
| 152 | IMMT | 13181 | -0.696 | -0.3099 | No | ||
| 153 | ZFYVE1 | 13277 | -0.709 | -0.3126 | No | ||
| 154 | MLLT7 | 13305 | -0.714 | -0.3115 | No | ||
| 155 | AP2M1 | 13311 | -0.715 | -0.3092 | No | ||
| 156 | HDAC8 | 13385 | -0.727 | -0.3106 | No | ||
| 157 | EXO1 | 13458 | -0.739 | -0.3119 | No | ||
| 158 | MRPL50 | 13507 | -0.752 | -0.3118 | No | ||
| 159 | DNAJB12 | 13609 | -0.770 | -0.3146 | No | ||
| 160 | THAP11 | 13619 | -0.772 | -0.3123 | No | ||
| 161 | RPL7 | 13743 | -0.794 | -0.3162 | No | ||
| 162 | CPEB3 | 13778 | -0.800 | -0.3152 | No | ||
| 163 | PPP6C | 13790 | -0.802 | -0.3129 | No | ||
| 164 | PGM2L1 | 13917 | -0.825 | -0.3169 | Yes | ||
| 165 | RDH14 | 13961 | -0.834 | -0.3162 | Yes | ||
| 166 | COX7B | 13987 | -0.839 | -0.3146 | Yes | ||
| 167 | TSC2 | 14028 | -0.849 | -0.3137 | Yes | ||
| 168 | MDH1 | 14032 | -0.849 | -0.3109 | Yes | ||
| 169 | CDC6 | 14058 | -0.854 | -0.3092 | Yes | ||
| 170 | ARID5A | 14174 | -0.877 | -0.3123 | Yes | ||
| 171 | CREBL1 | 14194 | -0.881 | -0.3102 | Yes | ||
| 172 | WBP2 | 14299 | -0.905 | -0.3127 | Yes | ||
| 173 | ASXL2 | 14315 | -0.907 | -0.3103 | Yes | ||
| 174 | TTC13 | 14364 | -0.916 | -0.3096 | Yes | ||
| 175 | PSMC4 | 14401 | -0.924 | -0.3083 | Yes | ||
| 176 | DNAJC12 | 14448 | -0.934 | -0.3075 | Yes | ||
| 177 | RPS27A | 14489 | -0.942 | -0.3063 | Yes | ||
| 178 | DNM1L | 14539 | -0.954 | -0.3055 | Yes | ||
| 179 | RBL2 | 14573 | -0.962 | -0.3039 | Yes | ||
| 180 | NUDT12 | 14611 | -0.971 | -0.3025 | Yes | ||
| 181 | SESN1 | 14636 | -0.976 | -0.3003 | Yes | ||
| 182 | VPS41 | 14699 | -0.988 | -0.3001 | Yes | ||
| 183 | ZDHHC14 | 14774 | -1.008 | -0.3006 | Yes | ||
| 184 | ADAM22 | 14782 | -1.011 | -0.2974 | Yes | ||
| 185 | PRIM1 | 14807 | -1.018 | -0.2950 | Yes | ||
| 186 | COL4A3BP | 14836 | -1.024 | -0.2929 | Yes | ||
| 187 | PPARGC1A | 14863 | -1.028 | -0.2907 | Yes | ||
| 188 | EIF2B4 | 14995 | -1.056 | -0.2940 | Yes | ||
| 189 | COMMD9 | 15126 | -1.089 | -0.2972 | Yes | ||
| 190 | VTI1B | 15237 | -1.121 | -0.2992 | Yes | ||
| 191 | RNASEH2A | 15246 | -1.124 | -0.2957 | Yes | ||
| 192 | HRSP12 | 15273 | -1.129 | -0.2931 | Yes | ||
| 193 | RABAC1 | 15423 | -1.167 | -0.2970 | Yes | ||
| 194 | UPF3B | 15545 | -1.201 | -0.2993 | Yes | ||
| 195 | BET1 | 15589 | -1.211 | -0.2973 | Yes | ||
| 196 | RALBP1 | 15601 | -1.214 | -0.2936 | Yes | ||
| 197 | RANGAP1 | 15606 | -1.215 | -0.2895 | Yes | ||
| 198 | WASF1 | 15624 | -1.218 | -0.2861 | Yes | ||
| 199 | SH3BGRL2 | 15644 | -1.222 | -0.2828 | Yes | ||
| 200 | FKBPL | 15734 | -1.249 | -0.2832 | Yes | ||
| 201 | FNTB | 15926 | -1.305 | -0.2889 | Yes | ||
| 202 | BACE1 | 15991 | -1.323 | -0.2877 | Yes | ||
| 203 | ADCK1 | 16049 | -1.342 | -0.2860 | Yes | ||
| 204 | ETFDH | 16067 | -1.348 | -0.2821 | Yes | ||
| 205 | TMOD3 | 16101 | -1.359 | -0.2791 | Yes | ||
| 206 | NTHL1 | 16291 | -1.425 | -0.2843 | Yes | ||
| 207 | RPL6 | 16318 | -1.435 | -0.2806 | Yes | ||
| 208 | SETMAR | 16382 | -1.459 | -0.2788 | Yes | ||
| 209 | OPHN1 | 16411 | -1.468 | -0.2751 | Yes | ||
| 210 | CRYZL1 | 16493 | -1.501 | -0.2742 | Yes | ||
| 211 | MAF1 | 16553 | -1.523 | -0.2720 | Yes | ||
| 212 | BANP | 16568 | -1.529 | -0.2673 | Yes | ||
| 213 | MRPL54 | 16590 | -1.538 | -0.2629 | Yes | ||
| 214 | KDELC1 | 16594 | -1.540 | -0.2576 | Yes | ||
| 215 | POLK | 16651 | -1.562 | -0.2551 | Yes | ||
| 216 | RUTBC3 | 16676 | -1.572 | -0.2508 | Yes | ||
| 217 | M6PR | 16728 | -1.596 | -0.2479 | Yes | ||
| 218 | KCNN3 | 16768 | -1.611 | -0.2443 | Yes | ||
| 219 | MRPL34 | 16798 | -1.623 | -0.2401 | Yes | ||
| 220 | TMEM4 | 16799 | -1.625 | -0.2343 | Yes | ||
| 221 | TNPO2 | 16801 | -1.625 | -0.2285 | Yes | ||
| 222 | NMT1 | 16871 | -1.659 | -0.2264 | Yes | ||
| 223 | RDH10 | 16899 | -1.675 | -0.2219 | Yes | ||
| 224 | ZNF228 | 16945 | -1.693 | -0.2183 | Yes | ||
| 225 | HBP1 | 16954 | -1.697 | -0.2127 | Yes | ||
| 226 | ZFP28 | 17056 | -1.752 | -0.2119 | Yes | ||
| 227 | PCM1 | 17128 | -1.796 | -0.2094 | Yes | ||
| 228 | PARK2 | 17175 | -1.816 | -0.2054 | Yes | ||
| 229 | PET112L | 17178 | -1.819 | -0.1991 | Yes | ||
| 230 | ABLIM2 | 17216 | -1.843 | -0.1945 | Yes | ||
| 231 | AAMP | 17411 | -1.958 | -0.1981 | Yes | ||
| 232 | ACBD5 | 17480 | -2.010 | -0.1946 | Yes | ||
| 233 | MRPL19 | 17525 | -2.041 | -0.1898 | Yes | ||
| 234 | SLC35B4 | 17558 | -2.068 | -0.1841 | Yes | ||
| 235 | LIG1 | 17601 | -2.111 | -0.1789 | Yes | ||
| 236 | UBE2H | 17683 | -2.162 | -0.1756 | Yes | ||
| 237 | ING3 | 17779 | -2.247 | -0.1728 | Yes | ||
| 238 | NOS3 | 17815 | -2.283 | -0.1665 | Yes | ||
| 239 | ZFP1 | 17842 | -2.310 | -0.1597 | Yes | ||
| 240 | PABPC1 | 17954 | -2.454 | -0.1570 | Yes | ||
| 241 | TRPC4AP | 18007 | -2.534 | -0.1508 | Yes | ||
| 242 | PDE6D | 18017 | -2.542 | -0.1423 | Yes | ||
| 243 | ACTR1A | 18026 | -2.553 | -0.1336 | Yes | ||
| 244 | CMAS | 18059 | -2.607 | -0.1260 | Yes | ||
| 245 | RNF121 | 18114 | -2.704 | -0.1193 | Yes | ||
| 246 | SKP2 | 18129 | -2.730 | -0.1104 | Yes | ||
| 247 | ABCC5 | 18147 | -2.750 | -0.1015 | Yes | ||
| 248 | HSPH1 | 18163 | -2.779 | -0.0924 | Yes | ||
| 249 | PRCC | 18240 | -2.954 | -0.0860 | Yes | ||
| 250 | SNX17 | 18256 | -2.991 | -0.0762 | Yes | ||
| 251 | FKBP4 | 18304 | -3.127 | -0.0676 | Yes | ||
| 252 | DCTN2 | 18323 | -3.195 | -0.0572 | Yes | ||
| 253 | FBXO9 | 18430 | -3.633 | -0.0500 | Yes | ||
| 254 | LGMN | 18527 | -4.564 | -0.0390 | Yes | ||
| 255 | LCMT1 | 18546 | -4.869 | -0.0226 | Yes | ||
| 256 | TP53 | 18597 | -7.392 | 0.0010 | Yes |