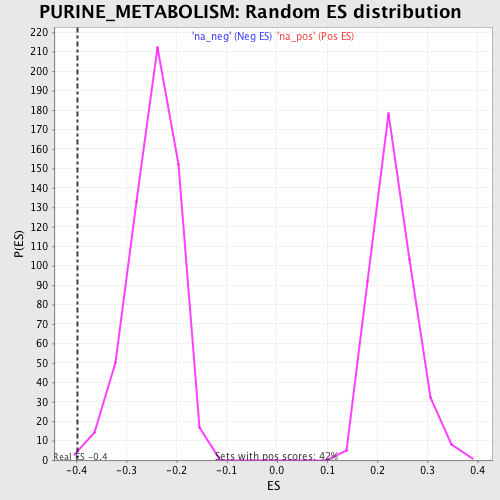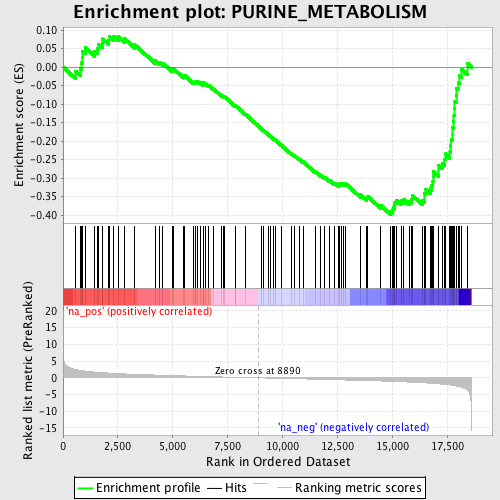
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PURINE_METABOLISM |
| Enrichment Score (ES) | -0.3983709 |
| Normalized Enrichment Score (NES) | -1.6337459 |
| Nominal p-value | 0.0051635113 |
| FDR q-value | 0.22373487 |
| FWER p-Value | 0.98 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PDE4B | 574 | 2.480 | -0.0107 | No | ||
| 2 | ADCY6 | 789 | 2.199 | -0.0042 | No | ||
| 3 | ADCY4 | 824 | 2.162 | 0.0117 | No | ||
| 4 | PDE4A | 883 | 2.109 | 0.0258 | No | ||
| 5 | GUCY1A3 | 890 | 2.099 | 0.0427 | No | ||
| 6 | GDA | 1015 | 1.997 | 0.0524 | No | ||
| 7 | AMPD3 | 1440 | 1.713 | 0.0435 | No | ||
| 8 | POLS | 1581 | 1.637 | 0.0494 | No | ||
| 9 | AK1 | 1606 | 1.625 | 0.0614 | No | ||
| 10 | RRM1 | 1788 | 1.537 | 0.0642 | No | ||
| 11 | POLR2J | 1803 | 1.531 | 0.0760 | No | ||
| 12 | PDE4C | 2072 | 1.419 | 0.0732 | No | ||
| 13 | AMPD1 | 2092 | 1.414 | 0.0838 | No | ||
| 14 | IMPDH1 | 2303 | 1.334 | 0.0834 | No | ||
| 15 | PDE7B | 2520 | 1.268 | 0.0821 | No | ||
| 16 | POLE | 2803 | 1.182 | 0.0765 | No | ||
| 17 | NT5E | 3269 | 1.052 | 0.0600 | No | ||
| 18 | ENPP1 | 4191 | 0.829 | 0.0171 | No | ||
| 19 | GUCY1B3 | 4396 | 0.781 | 0.0125 | No | ||
| 20 | ALLC | 4545 | 0.748 | 0.0106 | No | ||
| 21 | PDE9A | 4965 | 0.666 | -0.0066 | No | ||
| 22 | ADCY5 | 5034 | 0.654 | -0.0049 | No | ||
| 23 | POLR2I | 5491 | 0.567 | -0.0249 | No | ||
| 24 | POLR2C | 5512 | 0.562 | -0.0213 | No | ||
| 25 | FHIT | 5943 | 0.483 | -0.0406 | No | ||
| 26 | ENTPD1 | 6036 | 0.467 | -0.0417 | No | ||
| 27 | POLR2B | 6047 | 0.465 | -0.0385 | No | ||
| 28 | ECGF1 | 6120 | 0.452 | -0.0387 | No | ||
| 29 | POLR2E | 6254 | 0.428 | -0.0423 | No | ||
| 30 | GUCY2F | 6376 | 0.405 | -0.0456 | No | ||
| 31 | POLG | 6377 | 0.405 | -0.0422 | No | ||
| 32 | ADCY2 | 6488 | 0.385 | -0.0450 | No | ||
| 33 | GUCY2C | 6607 | 0.369 | -0.0484 | No | ||
| 34 | ATP5G2 | 6846 | 0.331 | -0.0585 | No | ||
| 35 | PDE6G | 7218 | 0.263 | -0.0764 | No | ||
| 36 | AK5 | 7306 | 0.249 | -0.0791 | No | ||
| 37 | POLR2G | 7365 | 0.239 | -0.0802 | No | ||
| 38 | PDE6B | 7862 | 0.163 | -0.1057 | No | ||
| 39 | ITPA | 7871 | 0.162 | -0.1048 | No | ||
| 40 | ATP1B1 | 8321 | 0.092 | -0.1283 | No | ||
| 41 | ADCY8 | 9027 | -0.021 | -0.1662 | No | ||
| 42 | ATP5H | 9119 | -0.034 | -0.1708 | No | ||
| 43 | PRPS2 | 9380 | -0.075 | -0.1843 | No | ||
| 44 | POLB | 9437 | -0.082 | -0.1866 | No | ||
| 45 | GART | 9580 | -0.102 | -0.1935 | No | ||
| 46 | POLR1B | 9661 | -0.113 | -0.1969 | No | ||
| 47 | ATP5G1 | 9931 | -0.157 | -0.2101 | No | ||
| 48 | ATP5A1 | 10391 | -0.222 | -0.2331 | No | ||
| 49 | APRT | 10540 | -0.248 | -0.2390 | No | ||
| 50 | ATP5F1 | 10760 | -0.286 | -0.2485 | No | ||
| 51 | GUCY1B2 | 10939 | -0.315 | -0.2556 | No | ||
| 52 | POLR2K | 11488 | -0.406 | -0.2818 | No | ||
| 53 | NT5C | 11738 | -0.446 | -0.2916 | No | ||
| 54 | IMPDH2 | 11932 | -0.476 | -0.2982 | No | ||
| 55 | PAICS | 12144 | -0.511 | -0.3054 | No | ||
| 56 | ADCY3 | 12388 | -0.554 | -0.3140 | No | ||
| 57 | NPR2 | 12543 | -0.584 | -0.3175 | No | ||
| 58 | NUDT2 | 12603 | -0.597 | -0.3158 | No | ||
| 59 | PDE6C | 12678 | -0.609 | -0.3148 | No | ||
| 60 | CANT1 | 12757 | -0.621 | -0.3139 | No | ||
| 61 | ENTPD2 | 12859 | -0.638 | -0.3141 | No | ||
| 62 | GUK1 | 13550 | -0.760 | -0.3452 | No | ||
| 63 | PAPSS2 | 13836 | -0.810 | -0.3539 | No | ||
| 64 | AMPD2 | 13865 | -0.815 | -0.3488 | No | ||
| 65 | POLRMT | 14475 | -0.939 | -0.3740 | No | ||
| 66 | POLR2A | 14928 | -1.043 | -0.3898 | Yes | ||
| 67 | PAPSS1 | 15021 | -1.062 | -0.3861 | Yes | ||
| 68 | GUCY1A2 | 15038 | -1.066 | -0.3782 | Yes | ||
| 69 | POLD2 | 15100 | -1.083 | -0.3726 | Yes | ||
| 70 | ATP5G3 | 15123 | -1.088 | -0.3649 | Yes | ||
| 71 | ATIC | 15192 | -1.107 | -0.3595 | Yes | ||
| 72 | POLR2H | 15400 | -1.161 | -0.3612 | Yes | ||
| 73 | ATP5C1 | 15531 | -1.198 | -0.3584 | Yes | ||
| 74 | PDE1A | 15790 | -1.265 | -0.3619 | Yes | ||
| 75 | POLQ | 15894 | -1.295 | -0.3569 | Yes | ||
| 76 | ATP5I | 15921 | -1.304 | -0.3476 | Yes | ||
| 77 | AK2 | 16372 | -1.453 | -0.3600 | Yes | ||
| 78 | ADSL | 16478 | -1.497 | -0.3534 | Yes | ||
| 79 | PDE8A | 16479 | -1.497 | -0.3411 | Yes | ||
| 80 | RRM2 | 16534 | -1.516 | -0.3316 | Yes | ||
| 81 | ATP5J2 | 16731 | -1.597 | -0.3291 | Yes | ||
| 82 | HPRT1 | 16796 | -1.623 | -0.3192 | Yes | ||
| 83 | ADCY7 | 16854 | -1.651 | -0.3088 | Yes | ||
| 84 | GMPS | 16866 | -1.658 | -0.2958 | Yes | ||
| 85 | ATP5B | 16881 | -1.666 | -0.2829 | Yes | ||
| 86 | ENPP3 | 17103 | -1.781 | -0.2802 | Yes | ||
| 87 | DGUOK | 17105 | -1.782 | -0.2657 | Yes | ||
| 88 | POLL | 17273 | -1.883 | -0.2592 | Yes | ||
| 89 | ATP5D | 17393 | -1.945 | -0.2497 | Yes | ||
| 90 | DCK | 17413 | -1.960 | -0.2347 | Yes | ||
| 91 | PDE5A | 17632 | -2.130 | -0.2290 | Yes | ||
| 92 | ADK | 17665 | -2.152 | -0.2131 | Yes | ||
| 93 | PPAT | 17679 | -2.158 | -0.1961 | Yes | ||
| 94 | PRPS1 | 17743 | -2.219 | -0.1813 | Yes | ||
| 95 | PKLR | 17746 | -2.220 | -0.1632 | Yes | ||
| 96 | NME2 | 17785 | -2.254 | -0.1468 | Yes | ||
| 97 | PKM2 | 17812 | -2.277 | -0.1295 | Yes | ||
| 98 | NT5M | 17846 | -2.313 | -0.1123 | Yes | ||
| 99 | POLD1 | 17858 | -2.338 | -0.0938 | Yes | ||
| 100 | NPR1 | 17913 | -2.398 | -0.0770 | Yes | ||
| 101 | PDE4D | 17934 | -2.425 | -0.0582 | Yes | ||
| 102 | NME1 | 18002 | -2.529 | -0.0411 | Yes | ||
| 103 | ADA | 18064 | -2.612 | -0.0230 | Yes | ||
| 104 | ATP5J | 18158 | -2.767 | -0.0053 | Yes | ||
| 105 | NP | 18437 | -3.659 | 0.0097 | Yes |