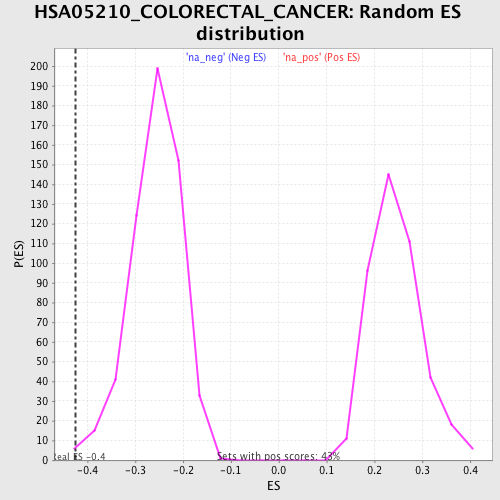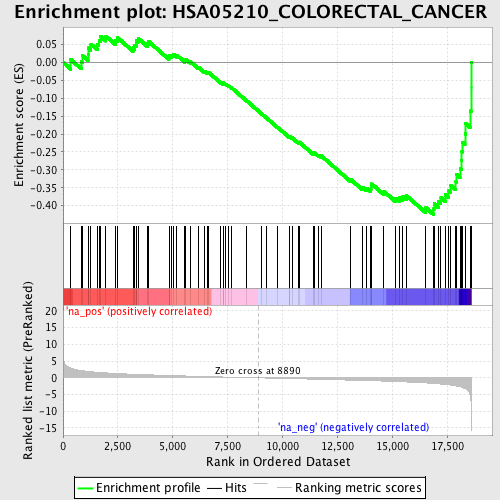
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA05210_COLORECTAL_CANCER |
| Enrichment Score (ES) | -0.42450887 |
| Normalized Enrichment Score (NES) | -1.6546649 |
| Nominal p-value | 0.007005254 |
| FDR q-value | 0.22526672 |
| FWER p-Value | 0.968 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCF7L2 | 348 | 2.868 | 0.0080 | No | ||
| 2 | RAF1 | 835 | 2.156 | 0.0019 | No | ||
| 3 | RAC2 | 867 | 2.129 | 0.0201 | No | ||
| 4 | AXIN2 | 1139 | 1.906 | 0.0233 | No | ||
| 5 | APC | 1149 | 1.899 | 0.0405 | No | ||
| 6 | PIK3CB | 1268 | 1.823 | 0.0512 | No | ||
| 7 | GRB2 | 1573 | 1.641 | 0.0501 | No | ||
| 8 | SMAD3 | 1635 | 1.611 | 0.0619 | No | ||
| 9 | BIRC5 | 1706 | 1.575 | 0.0728 | No | ||
| 10 | IGF1R | 1948 | 1.469 | 0.0735 | No | ||
| 11 | TGFBR1 | 2378 | 1.313 | 0.0626 | No | ||
| 12 | RALGDS | 2481 | 1.279 | 0.0691 | No | ||
| 13 | SMAD2 | 3188 | 1.075 | 0.0410 | No | ||
| 14 | BCL2 | 3260 | 1.054 | 0.0470 | No | ||
| 15 | MYC | 3332 | 1.038 | 0.0529 | No | ||
| 16 | MAPK10 | 3350 | 1.032 | 0.0616 | No | ||
| 17 | FZD5 | 3437 | 1.009 | 0.0664 | No | ||
| 18 | PIK3R5 | 3860 | 0.902 | 0.0520 | No | ||
| 19 | TCF7 | 3887 | 0.897 | 0.0590 | No | ||
| 20 | SOS1 | 4827 | 0.690 | 0.0148 | No | ||
| 21 | EGFR | 4843 | 0.688 | 0.0204 | No | ||
| 22 | PIK3CG | 4959 | 0.667 | 0.0204 | No | ||
| 23 | PDGFRB | 5027 | 0.654 | 0.0229 | No | ||
| 24 | DVL2 | 5185 | 0.623 | 0.0203 | No | ||
| 25 | FZD8 | 5553 | 0.555 | 0.0056 | No | ||
| 26 | BAD | 5594 | 0.547 | 0.0086 | No | ||
| 27 | FZD9 | 5810 | 0.507 | 0.0017 | No | ||
| 28 | MSH3 | 6163 | 0.445 | -0.0131 | No | ||
| 29 | APC2 | 6435 | 0.395 | -0.0240 | No | ||
| 30 | GSK3B | 6584 | 0.372 | -0.0286 | No | ||
| 31 | PIK3R1 | 6623 | 0.366 | -0.0272 | No | ||
| 32 | AKT1 | 7194 | 0.267 | -0.0554 | No | ||
| 33 | PIK3R2 | 7294 | 0.252 | -0.0584 | No | ||
| 34 | CCND1 | 7311 | 0.248 | -0.0570 | No | ||
| 35 | PIK3CA | 7419 | 0.232 | -0.0606 | No | ||
| 36 | MET | 7521 | 0.217 | -0.0640 | No | ||
| 37 | PDGFRA | 7692 | 0.189 | -0.0714 | No | ||
| 38 | FZD1 | 8380 | 0.083 | -0.1077 | No | ||
| 39 | FZD4 | 9029 | -0.021 | -0.1425 | No | ||
| 40 | TGFB2 | 9260 | -0.055 | -0.1543 | No | ||
| 41 | RAC1 | 9785 | -0.132 | -0.1814 | No | ||
| 42 | TGFB1 | 10324 | -0.214 | -0.2084 | No | ||
| 43 | SOS2 | 10339 | -0.215 | -0.2072 | No | ||
| 44 | PIK3R3 | 10441 | -0.230 | -0.2105 | No | ||
| 45 | AXIN1 | 10731 | -0.281 | -0.2234 | No | ||
| 46 | MAP2K1 | 10757 | -0.285 | -0.2221 | No | ||
| 47 | MAPK3 | 11428 | -0.397 | -0.2545 | No | ||
| 48 | FZD10 | 11444 | -0.399 | -0.2516 | No | ||
| 49 | MLH1 | 11650 | -0.432 | -0.2587 | No | ||
| 50 | FZD7 | 11771 | -0.451 | -0.2609 | No | ||
| 51 | TGFBR2 | 13088 | -0.682 | -0.3255 | No | ||
| 52 | FZD2 | 13647 | -0.777 | -0.3484 | No | ||
| 53 | FZD3 | 13841 | -0.811 | -0.3512 | No | ||
| 54 | RAC3 | 14030 | -0.849 | -0.3535 | No | ||
| 55 | DVL3 | 14034 | -0.849 | -0.3457 | No | ||
| 56 | MSH6 | 14063 | -0.855 | -0.3392 | No | ||
| 57 | CYCS | 14621 | -0.973 | -0.3602 | No | ||
| 58 | DVL1 | 15154 | -1.097 | -0.3786 | No | ||
| 59 | CASP9 | 15313 | -1.138 | -0.3765 | No | ||
| 60 | TGFB3 | 15487 | -1.185 | -0.3748 | No | ||
| 61 | KRAS | 15643 | -1.222 | -0.3718 | No | ||
| 62 | AKT3 | 16533 | -1.515 | -0.4056 | No | ||
| 63 | FOS | 16885 | -1.668 | -0.4089 | Yes | ||
| 64 | SMAD4 | 16907 | -1.677 | -0.3944 | Yes | ||
| 65 | ARAF | 17122 | -1.791 | -0.3892 | Yes | ||
| 66 | ACVR1C | 17221 | -1.849 | -0.3773 | Yes | ||
| 67 | MAPK8 | 17416 | -1.961 | -0.3694 | Yes | ||
| 68 | MAPK9 | 17583 | -2.095 | -0.3588 | Yes | ||
| 69 | MSH2 | 17672 | -2.155 | -0.3434 | Yes | ||
| 70 | CASP3 | 17902 | -2.386 | -0.3335 | Yes | ||
| 71 | FZD6 | 17916 | -2.402 | -0.3118 | Yes | ||
| 72 | AKT2 | 18092 | -2.672 | -0.2963 | Yes | ||
| 73 | LEF1 | 18162 | -2.778 | -0.2741 | Yes | ||
| 74 | DCC | 18176 | -2.820 | -0.2485 | Yes | ||
| 75 | MAPK1 | 18210 | -2.885 | -0.2233 | Yes | ||
| 76 | PIK3CD | 18319 | -3.174 | -0.1995 | Yes | ||
| 77 | JUN | 18337 | -3.236 | -0.1702 | Yes | ||
| 78 | ACVR1B | 18550 | -4.949 | -0.1354 | Yes | ||
| 79 | TP53 | 18597 | -7.392 | -0.0689 | Yes | ||
| 80 | BAX | 18598 | -7.485 | 0.0010 | Yes |