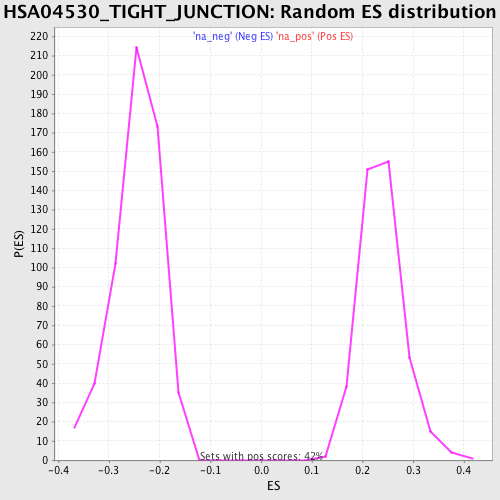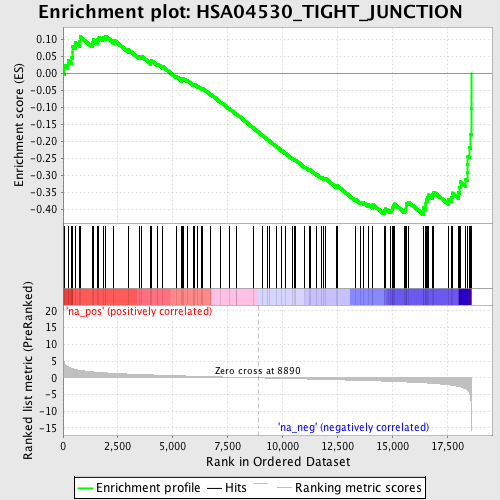
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04530_TIGHT_JUNCTION |
| Enrichment Score (ES) | -0.4147397 |
| Normalized Enrichment Score (NES) | -1.6959724 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.25525165 |
| FWER p-Value | 0.912 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MYL9 | 77 | 4.222 | 0.0236 | No | ||
| 2 | MYL2 | 222 | 3.278 | 0.0374 | No | ||
| 3 | PRKCD | 368 | 2.828 | 0.0482 | No | ||
| 4 | CLDN15 | 439 | 2.698 | 0.0622 | No | ||
| 5 | TJP3 | 442 | 2.693 | 0.0798 | No | ||
| 6 | CLDN1 | 551 | 2.518 | 0.0905 | No | ||
| 7 | ASH1L | 763 | 2.224 | 0.0937 | No | ||
| 8 | RRAS | 770 | 2.216 | 0.1080 | No | ||
| 9 | PRKCB1 | 1323 | 1.788 | 0.0900 | No | ||
| 10 | CLDN5 | 1373 | 1.751 | 0.0988 | No | ||
| 11 | MYH11 | 1554 | 1.650 | 0.1000 | No | ||
| 12 | CLDN7 | 1633 | 1.611 | 0.1064 | No | ||
| 13 | MYL7 | 1818 | 1.527 | 0.1065 | No | ||
| 14 | MYLPF | 1936 | 1.474 | 0.1099 | No | ||
| 15 | JAM2 | 2317 | 1.329 | 0.0981 | No | ||
| 16 | PRKCE | 2978 | 1.130 | 0.0699 | No | ||
| 17 | CLDN8 | 3467 | 0.999 | 0.0501 | No | ||
| 18 | MYH9 | 3594 | 0.965 | 0.0497 | No | ||
| 19 | CLDN11 | 4004 | 0.874 | 0.0333 | No | ||
| 20 | CRB3 | 4033 | 0.866 | 0.0375 | No | ||
| 21 | PPP2R4 | 4317 | 0.795 | 0.0275 | No | ||
| 22 | CSNK2A1 | 4538 | 0.749 | 0.0205 | No | ||
| 23 | ACTN4 | 5162 | 0.627 | -0.0090 | No | ||
| 24 | CASK | 5415 | 0.581 | -0.0188 | No | ||
| 25 | CLDN23 | 5434 | 0.577 | -0.0160 | No | ||
| 26 | PRKCZ | 5502 | 0.564 | -0.0159 | No | ||
| 27 | JAM3 | 5652 | 0.539 | -0.0204 | No | ||
| 28 | EXOC3 | 5952 | 0.483 | -0.0333 | No | ||
| 29 | CGN | 5969 | 0.479 | -0.0311 | No | ||
| 30 | CLDN19 | 6138 | 0.449 | -0.0372 | No | ||
| 31 | CLDN9 | 6302 | 0.417 | -0.0432 | No | ||
| 32 | MRAS | 6372 | 0.406 | -0.0443 | No | ||
| 33 | ACTG1 | 6729 | 0.351 | -0.0612 | No | ||
| 34 | AKT1 | 7194 | 0.267 | -0.0845 | No | ||
| 35 | VAPA | 7587 | 0.206 | -0.1043 | No | ||
| 36 | CLDN2 | 7908 | 0.155 | -0.1206 | No | ||
| 37 | MYH3 | 7922 | 0.153 | -0.1203 | No | ||
| 38 | PTEN | 8691 | 0.035 | -0.1615 | No | ||
| 39 | YES1 | 9079 | -0.029 | -0.1822 | No | ||
| 40 | GNAI1 | 9326 | -0.066 | -0.1951 | No | ||
| 41 | RAB13 | 9410 | -0.079 | -0.1990 | No | ||
| 42 | PPP2R1A | 9721 | -0.123 | -0.2150 | No | ||
| 43 | NRAS | 9973 | -0.163 | -0.2274 | No | ||
| 44 | CSNK2B | 10120 | -0.184 | -0.2341 | No | ||
| 45 | PARD6G | 10472 | -0.236 | -0.2515 | No | ||
| 46 | PRKCI | 10527 | -0.246 | -0.2528 | No | ||
| 47 | CTTN | 10612 | -0.260 | -0.2556 | No | ||
| 48 | PPP2CB | 11014 | -0.328 | -0.2751 | No | ||
| 49 | PARD6B | 11225 | -0.365 | -0.2841 | No | ||
| 50 | PRKCQ | 11256 | -0.370 | -0.2832 | No | ||
| 51 | CLDN18 | 11569 | -0.419 | -0.2973 | No | ||
| 52 | MYH4 | 11785 | -0.453 | -0.3060 | No | ||
| 53 | PPP2CA | 11880 | -0.467 | -0.3080 | No | ||
| 54 | MPDZ | 11938 | -0.476 | -0.3079 | No | ||
| 55 | MYH6 | 12475 | -0.571 | -0.3331 | No | ||
| 56 | MYH2 | 12489 | -0.575 | -0.3300 | No | ||
| 57 | MYH7 | 13325 | -0.717 | -0.3704 | No | ||
| 58 | CLDN4 | 13565 | -0.762 | -0.3783 | No | ||
| 59 | ACTN3 | 13692 | -0.784 | -0.3799 | No | ||
| 60 | PARD6A | 13897 | -0.822 | -0.3855 | No | ||
| 61 | PARD3 | 14104 | -0.863 | -0.3910 | No | ||
| 62 | RAB3B | 14121 | -0.866 | -0.3861 | No | ||
| 63 | MYH14 | 14626 | -0.974 | -0.4069 | Yes | ||
| 64 | TJP1 | 14653 | -0.979 | -0.4019 | Yes | ||
| 65 | RHOA | 14709 | -0.991 | -0.3984 | Yes | ||
| 66 | CLDN17 | 14932 | -1.043 | -0.4035 | Yes | ||
| 67 | SRC | 14989 | -1.055 | -0.3996 | Yes | ||
| 68 | HRAS | 15010 | -1.058 | -0.3937 | Yes | ||
| 69 | CLDN10 | 15043 | -1.068 | -0.3884 | Yes | ||
| 70 | PPP2R2A | 15090 | -1.079 | -0.3838 | Yes | ||
| 71 | PPM1J | 15576 | -1.208 | -0.4020 | Yes | ||
| 72 | CLDN14 | 15627 | -1.218 | -0.3967 | Yes | ||
| 73 | GNAI3 | 15629 | -1.219 | -0.3887 | Yes | ||
| 74 | KRAS | 15643 | -1.222 | -0.3814 | Yes | ||
| 75 | MPP5 | 15735 | -1.249 | -0.3781 | Yes | ||
| 76 | OCLN | 16415 | -1.472 | -0.4051 | Yes | ||
| 77 | HCLS1 | 16418 | -1.473 | -0.3955 | Yes | ||
| 78 | F11R | 16530 | -1.515 | -0.3915 | Yes | ||
| 79 | AKT3 | 16533 | -1.515 | -0.3816 | Yes | ||
| 80 | PRKCA | 16539 | -1.517 | -0.3719 | Yes | ||
| 81 | CLDN16 | 16616 | -1.549 | -0.3658 | Yes | ||
| 82 | CLDN3 | 16650 | -1.561 | -0.3573 | Yes | ||
| 83 | ACTN2 | 16842 | -1.646 | -0.3568 | Yes | ||
| 84 | RRAS2 | 16898 | -1.674 | -0.3488 | Yes | ||
| 85 | CSNK2A2 | 17557 | -2.068 | -0.3707 | Yes | ||
| 86 | CDC42 | 17688 | -2.168 | -0.3634 | Yes | ||
| 87 | CSDA | 17738 | -2.212 | -0.3515 | Yes | ||
| 88 | TJP2 | 18029 | -2.555 | -0.3504 | Yes | ||
| 89 | ACTB | 18073 | -2.633 | -0.3354 | Yes | ||
| 90 | AKT2 | 18092 | -2.672 | -0.3188 | Yes | ||
| 91 | CDK4 | 18355 | -3.310 | -0.3111 | Yes | ||
| 92 | MAGI1 | 18422 | -3.588 | -0.2911 | Yes | ||
| 93 | MYH10 | 18433 | -3.640 | -0.2677 | Yes | ||
| 94 | INADL | 18450 | -3.746 | -0.2439 | Yes | ||
| 95 | EXOC4 | 18535 | -4.671 | -0.2177 | Yes | ||
| 96 | PRKCH | 18586 | -6.190 | -0.1797 | Yes | ||
| 97 | MYLC2PL | 18612 | -11.925 | -0.1025 | Yes | ||
| 98 | AMOTL1 | 18616 | -15.608 | 0.0000 | Yes |