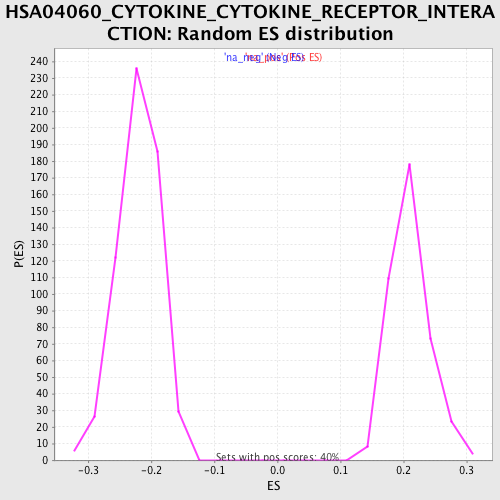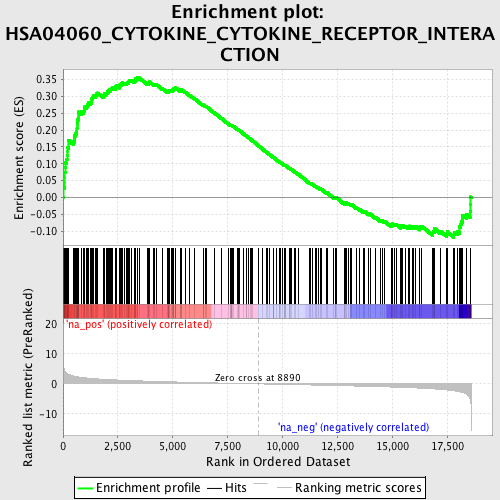
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04060_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION |
| Enrichment Score (ES) | 0.35525003 |
| Normalized Enrichment Score (NES) | 1.6957132 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.19858564 |
| FWER p-Value | 0.912 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KIT | 7 | 8.558 | 0.0312 | Yes | ||
| 2 | PRLR | 68 | 4.349 | 0.0439 | Yes | ||
| 3 | IL21R | 70 | 4.336 | 0.0598 | Yes | ||
| 4 | IL9R | 73 | 4.303 | 0.0756 | Yes | ||
| 5 | CXCL12 | 99 | 4.042 | 0.0891 | Yes | ||
| 6 | TNFSF13B | 109 | 3.947 | 0.1032 | Yes | ||
| 7 | CSF3R | 164 | 3.546 | 0.1133 | Yes | ||
| 8 | IL5 | 184 | 3.466 | 0.1250 | Yes | ||
| 9 | LTBR | 205 | 3.349 | 0.1363 | Yes | ||
| 10 | LTB | 209 | 3.330 | 0.1484 | Yes | ||
| 11 | CX3CR1 | 254 | 3.142 | 0.1576 | Yes | ||
| 12 | IL8RB | 255 | 3.140 | 0.1692 | Yes | ||
| 13 | CLCF1 | 466 | 2.648 | 0.1675 | Yes | ||
| 14 | IL1B | 525 | 2.550 | 0.1737 | Yes | ||
| 15 | TNFSF14 | 534 | 2.539 | 0.1827 | Yes | ||
| 16 | EPOR | 562 | 2.497 | 0.1904 | Yes | ||
| 17 | IL10RA | 626 | 2.414 | 0.1959 | Yes | ||
| 18 | TNFRSF13C | 631 | 2.410 | 0.2045 | Yes | ||
| 19 | CNTF | 637 | 2.405 | 0.2131 | Yes | ||
| 20 | IFNA4 | 641 | 2.400 | 0.2218 | Yes | ||
| 21 | CSF1R | 652 | 2.382 | 0.2300 | Yes | ||
| 22 | FLT1 | 685 | 2.327 | 0.2369 | Yes | ||
| 23 | CD40 | 692 | 2.315 | 0.2451 | Yes | ||
| 24 | TNFRSF1A | 694 | 2.313 | 0.2535 | Yes | ||
| 25 | TNFRSF1B | 830 | 2.158 | 0.2542 | Yes | ||
| 26 | VEGFA | 934 | 2.062 | 0.2562 | Yes | ||
| 27 | IL21 | 957 | 2.048 | 0.2625 | Yes | ||
| 28 | CCL27 | 988 | 2.019 | 0.2683 | Yes | ||
| 29 | IL6ST | 1083 | 1.941 | 0.2704 | Yes | ||
| 30 | IL7 | 1125 | 1.916 | 0.2752 | Yes | ||
| 31 | IFNGR1 | 1157 | 1.896 | 0.2805 | Yes | ||
| 32 | TNFRSF12A | 1247 | 1.835 | 0.2824 | Yes | ||
| 33 | EGF | 1302 | 1.804 | 0.2861 | Yes | ||
| 34 | CXCL2 | 1311 | 1.800 | 0.2923 | Yes | ||
| 35 | TNFSF13 | 1334 | 1.779 | 0.2977 | Yes | ||
| 36 | CSF2RA | 1385 | 1.745 | 0.3014 | Yes | ||
| 37 | IFNA7 | 1478 | 1.687 | 0.3026 | Yes | ||
| 38 | TNFRSF18 | 1514 | 1.670 | 0.3069 | Yes | ||
| 39 | TNFRSF11B | 1575 | 1.640 | 0.3096 | Yes | ||
| 40 | IL1RAP | 1831 | 1.520 | 0.3014 | Yes | ||
| 41 | TNFRSF25 | 1894 | 1.491 | 0.3035 | Yes | ||
| 42 | TNFRSF8 | 1900 | 1.488 | 0.3087 | Yes | ||
| 43 | CXCL9 | 1995 | 1.453 | 0.3090 | Yes | ||
| 44 | GDF5 | 2012 | 1.448 | 0.3134 | Yes | ||
| 45 | CCR7 | 2066 | 1.421 | 0.3158 | Yes | ||
| 46 | TNFRSF9 | 2105 | 1.408 | 0.3189 | Yes | ||
| 47 | IL1R2 | 2170 | 1.388 | 0.3205 | Yes | ||
| 48 | IL23R | 2195 | 1.375 | 0.3243 | Yes | ||
| 49 | IL1R1 | 2252 | 1.353 | 0.3263 | Yes | ||
| 50 | TGFBR1 | 2378 | 1.313 | 0.3243 | Yes | ||
| 51 | XCR1 | 2395 | 1.307 | 0.3282 | Yes | ||
| 52 | IL24 | 2417 | 1.303 | 0.3319 | Yes | ||
| 53 | IL22RA2 | 2559 | 1.255 | 0.3289 | Yes | ||
| 54 | IL17RA | 2563 | 1.254 | 0.3333 | Yes | ||
| 55 | IL23A | 2622 | 1.235 | 0.3347 | Yes | ||
| 56 | CNTFR | 2654 | 1.223 | 0.3375 | Yes | ||
| 57 | PF4 | 2684 | 1.215 | 0.3404 | Yes | ||
| 58 | CCL19 | 2808 | 1.180 | 0.3381 | Yes | ||
| 59 | TNFRSF10B | 2876 | 1.159 | 0.3387 | Yes | ||
| 60 | AMHR2 | 2917 | 1.148 | 0.3408 | Yes | ||
| 61 | IL17B | 2990 | 1.127 | 0.3410 | Yes | ||
| 62 | TNF | 2999 | 1.123 | 0.3447 | Yes | ||
| 63 | IL2RG | 3041 | 1.112 | 0.3466 | Yes | ||
| 64 | CD40LG | 3108 | 1.097 | 0.3471 | Yes | ||
| 65 | TNFRSF4 | 3248 | 1.057 | 0.3434 | Yes | ||
| 66 | CCL24 | 3256 | 1.055 | 0.3469 | Yes | ||
| 67 | IFNK | 3298 | 1.045 | 0.3485 | Yes | ||
| 68 | IL28RA | 3304 | 1.044 | 0.3521 | Yes | ||
| 69 | ACVR2A | 3379 | 1.024 | 0.3519 | Yes | ||
| 70 | CXCL14 | 3387 | 1.023 | 0.3553 | Yes | ||
| 71 | TNFSF15 | 3475 | 0.997 | 0.3542 | No | ||
| 72 | IL12A | 3838 | 0.907 | 0.3379 | No | ||
| 73 | IL9 | 3880 | 0.899 | 0.3389 | No | ||
| 74 | IL15RA | 3911 | 0.892 | 0.3406 | No | ||
| 75 | CXCL10 | 3936 | 0.888 | 0.3426 | No | ||
| 76 | IL18RAP | 4127 | 0.845 | 0.3354 | No | ||
| 77 | INHBC | 4187 | 0.831 | 0.3352 | No | ||
| 78 | PPBP | 4243 | 0.814 | 0.3352 | No | ||
| 79 | IL12RB1 | 4512 | 0.753 | 0.3234 | No | ||
| 80 | GHR | 4748 | 0.707 | 0.3133 | No | ||
| 81 | EPO | 4808 | 0.694 | 0.3126 | No | ||
| 82 | IFNB1 | 4818 | 0.692 | 0.3147 | No | ||
| 83 | BMPR1A | 4825 | 0.691 | 0.3169 | No | ||
| 84 | EGFR | 4843 | 0.688 | 0.3185 | No | ||
| 85 | IL19 | 4948 | 0.669 | 0.3153 | No | ||
| 86 | IL11 | 4972 | 0.664 | 0.3165 | No | ||
| 87 | CCR1 | 4977 | 0.663 | 0.3187 | No | ||
| 88 | CCL4 | 5023 | 0.655 | 0.3187 | No | ||
| 89 | PDGFRB | 5027 | 0.654 | 0.3210 | No | ||
| 90 | IL6R | 5051 | 0.650 | 0.3221 | No | ||
| 91 | KITLG | 5101 | 0.637 | 0.3218 | No | ||
| 92 | CD27 | 5114 | 0.636 | 0.3235 | No | ||
| 93 | TNFRSF21 | 5131 | 0.632 | 0.3249 | No | ||
| 94 | LIF | 5329 | 0.596 | 0.3164 | No | ||
| 95 | IL13RA1 | 5371 | 0.588 | 0.3164 | No | ||
| 96 | CCL8 | 5374 | 0.588 | 0.3184 | No | ||
| 97 | IL4R | 5376 | 0.587 | 0.3205 | No | ||
| 98 | IL12RB2 | 5568 | 0.552 | 0.3122 | No | ||
| 99 | AMH | 5781 | 0.512 | 0.3026 | No | ||
| 100 | LTA | 5983 | 0.476 | 0.2934 | No | ||
| 101 | BMPR2 | 6381 | 0.405 | 0.2733 | No | ||
| 102 | LIFR | 6390 | 0.402 | 0.2743 | No | ||
| 103 | IL20 | 6482 | 0.386 | 0.2708 | No | ||
| 104 | CCR3 | 6522 | 0.380 | 0.2701 | No | ||
| 105 | FLT4 | 6885 | 0.325 | 0.2516 | No | ||
| 106 | CCL22 | 7207 | 0.265 | 0.2352 | No | ||
| 107 | MET | 7521 | 0.217 | 0.2189 | No | ||
| 108 | IL25 | 7626 | 0.200 | 0.2140 | No | ||
| 109 | CD70 | 7632 | 0.199 | 0.2145 | No | ||
| 110 | EDA2R | 7679 | 0.192 | 0.2127 | No | ||
| 111 | CCR6 | 7688 | 0.190 | 0.2130 | No | ||
| 112 | PDGFRA | 7692 | 0.189 | 0.2135 | No | ||
| 113 | CCR4 | 7712 | 0.186 | 0.2132 | No | ||
| 114 | BMP2 | 7764 | 0.179 | 0.2110 | No | ||
| 115 | IFNA14 | 7929 | 0.151 | 0.2027 | No | ||
| 116 | CXCL1 | 7985 | 0.141 | 0.2002 | No | ||
| 117 | CXCL5 | 8008 | 0.137 | 0.1995 | No | ||
| 118 | IL22RA1 | 8047 | 0.130 | 0.1979 | No | ||
| 119 | CSF1 | 8221 | 0.106 | 0.1889 | No | ||
| 120 | CCL2 | 8360 | 0.085 | 0.1817 | No | ||
| 121 | CCR8 | 8375 | 0.083 | 0.1813 | No | ||
| 122 | CXCR4 | 8439 | 0.072 | 0.1781 | No | ||
| 123 | CCL17 | 8534 | 0.057 | 0.1732 | No | ||
| 124 | IFNAR1 | 8579 | 0.051 | 0.1710 | No | ||
| 125 | IL6 | 8650 | 0.040 | 0.1674 | No | ||
| 126 | CTF1 | 8892 | -0.000 | 0.1543 | No | ||
| 127 | IL2RA | 9084 | -0.029 | 0.1440 | No | ||
| 128 | TGFB2 | 9260 | -0.055 | 0.1347 | No | ||
| 129 | CXCL11 | 9327 | -0.066 | 0.1313 | No | ||
| 130 | HGF | 9413 | -0.079 | 0.1270 | No | ||
| 131 | TNFRSF17 | 9575 | -0.101 | 0.1186 | No | ||
| 132 | CCL11 | 9591 | -0.103 | 0.1182 | No | ||
| 133 | ACVR2B | 9606 | -0.105 | 0.1178 | No | ||
| 134 | VEGFC | 9703 | -0.119 | 0.1131 | No | ||
| 135 | IL8RA | 9872 | -0.147 | 0.1045 | No | ||
| 136 | IL2 | 9880 | -0.148 | 0.1046 | No | ||
| 137 | IFNA13 | 9888 | -0.149 | 0.1048 | No | ||
| 138 | IL12B | 9978 | -0.163 | 0.1006 | No | ||
| 139 | CCR9 | 10081 | -0.179 | 0.0957 | No | ||
| 140 | TNFRSF14 | 10105 | -0.181 | 0.0951 | No | ||
| 141 | IFNA6 | 10124 | -0.184 | 0.0948 | No | ||
| 142 | IFNA1 | 10137 | -0.186 | 0.0948 | No | ||
| 143 | MPL | 10302 | -0.210 | 0.0867 | No | ||
| 144 | TGFB1 | 10324 | -0.214 | 0.0863 | No | ||
| 145 | IL18R1 | 10359 | -0.218 | 0.0853 | No | ||
| 146 | TPO | 10404 | -0.223 | 0.0837 | No | ||
| 147 | OSMR | 10559 | -0.251 | 0.0763 | No | ||
| 148 | IL4 | 10599 | -0.259 | 0.0751 | No | ||
| 149 | EDA | 10736 | -0.282 | 0.0688 | No | ||
| 150 | TNFSF18 | 11246 | -0.369 | 0.0425 | No | ||
| 151 | IL3RA | 11266 | -0.372 | 0.0428 | No | ||
| 152 | CCL3 | 11345 | -0.384 | 0.0400 | No | ||
| 153 | IFNA2 | 11485 | -0.405 | 0.0339 | No | ||
| 154 | CRLF2 | 11560 | -0.417 | 0.0314 | No | ||
| 155 | BMP7 | 11637 | -0.430 | 0.0289 | No | ||
| 156 | IFNGR2 | 11712 | -0.441 | 0.0265 | No | ||
| 157 | XCL1 | 11781 | -0.452 | 0.0245 | No | ||
| 158 | BMPR1B | 11993 | -0.485 | 0.0148 | No | ||
| 159 | CSF2 | 12036 | -0.492 | 0.0143 | No | ||
| 160 | IL18 | 12335 | -0.546 | 0.0001 | No | ||
| 161 | TNFRSF13B | 12392 | -0.554 | -0.0009 | No | ||
| 162 | PRL | 12434 | -0.561 | -0.0010 | No | ||
| 163 | IFNE1 | 12442 | -0.562 | 0.0006 | No | ||
| 164 | PDGFB | 12824 | -0.633 | -0.0177 | No | ||
| 165 | EDAR | 12844 | -0.635 | -0.0164 | No | ||
| 166 | FLT3 | 12867 | -0.640 | -0.0153 | No | ||
| 167 | PDGFC | 12899 | -0.647 | -0.0146 | No | ||
| 168 | IL1A | 12999 | -0.667 | -0.0175 | No | ||
| 169 | TGFBR2 | 13088 | -0.682 | -0.0198 | No | ||
| 170 | IL22 | 13156 | -0.692 | -0.0208 | No | ||
| 171 | IL13 | 13353 | -0.721 | -0.0288 | No | ||
| 172 | TNFRSF11A | 13529 | -0.757 | -0.0356 | No | ||
| 173 | CXCR3 | 13707 | -0.788 | -0.0423 | No | ||
| 174 | LEP | 13748 | -0.795 | -0.0415 | No | ||
| 175 | NGFR | 13932 | -0.829 | -0.0484 | No | ||
| 176 | TNFSF12 | 13989 | -0.840 | -0.0484 | No | ||
| 177 | CX3CL1 | 14219 | -0.886 | -0.0575 | No | ||
| 178 | IFNAR2 | 14474 | -0.938 | -0.0679 | No | ||
| 179 | IL5RA | 14552 | -0.958 | -0.0685 | No | ||
| 180 | CCL28 | 14642 | -0.978 | -0.0698 | No | ||
| 181 | CCL25 | 14959 | -1.047 | -0.0831 | No | ||
| 182 | TNFSF9 | 14971 | -1.051 | -0.0798 | No | ||
| 183 | IFNG | 14994 | -1.056 | -0.0771 | No | ||
| 184 | IL3 | 15103 | -1.084 | -0.0790 | No | ||
| 185 | VEGFB | 15183 | -1.105 | -0.0792 | No | ||
| 186 | CCL1 | 15387 | -1.158 | -0.0860 | No | ||
| 187 | KDR | 15406 | -1.163 | -0.0827 | No | ||
| 188 | TGFB3 | 15487 | -1.185 | -0.0827 | No | ||
| 189 | CXCL13 | 15602 | -1.214 | -0.0844 | No | ||
| 190 | CXCR6 | 15744 | -1.250 | -0.0874 | No | ||
| 191 | TNFSF4 | 15767 | -1.257 | -0.0840 | No | ||
| 192 | INHBE | 15924 | -1.304 | -0.0877 | No | ||
| 193 | CCL7 | 15949 | -1.311 | -0.0842 | No | ||
| 194 | IL17RB | 16064 | -1.347 | -0.0854 | No | ||
| 195 | CCR5 | 16258 | -1.412 | -0.0907 | No | ||
| 196 | CSF3 | 16264 | -1.413 | -0.0857 | No | ||
| 197 | FASLG | 16350 | -1.445 | -0.0850 | No | ||
| 198 | IFNA5 | 16855 | -1.651 | -0.1063 | No | ||
| 199 | IL2RB | 16858 | -1.652 | -0.1004 | No | ||
| 200 | CCR2 | 16927 | -1.684 | -0.0979 | No | ||
| 201 | LEPR | 16942 | -1.691 | -0.0924 | No | ||
| 202 | TNFSF8 | 17198 | -1.832 | -0.0995 | No | ||
| 203 | FAS | 17486 | -2.013 | -0.1077 | No | ||
| 204 | CXCL16 | 17500 | -2.023 | -0.1009 | No | ||
| 205 | INHBB | 17814 | -2.278 | -0.1095 | No | ||
| 206 | IL10RB | 17835 | -2.303 | -0.1021 | No | ||
| 207 | TNFRSF19 | 17972 | -2.483 | -0.1004 | No | ||
| 208 | IL7R | 18074 | -2.641 | -0.0961 | No | ||
| 209 | IL10 | 18075 | -2.641 | -0.0864 | No | ||
| 210 | TSLP | 18123 | -2.724 | -0.0789 | No | ||
| 211 | CCL5 | 18148 | -2.750 | -0.0701 | No | ||
| 212 | ACVR1 | 18200 | -2.870 | -0.0623 | No | ||
| 213 | TNFSF11 | 18214 | -2.891 | -0.0523 | No | ||
| 214 | IL17A | 18387 | -3.424 | -0.0491 | No | ||
| 215 | ACVR1B | 18550 | -4.949 | -0.0397 | No | ||
| 216 | TNFSF10 | 18575 | -5.771 | -0.0197 | No | ||
| 217 | IL15 | 18580 | -5.934 | 0.0020 | No |