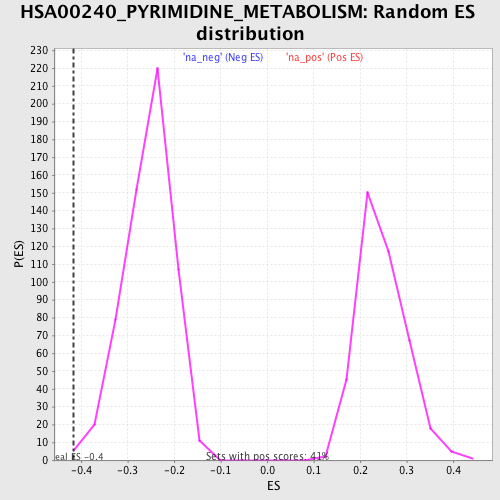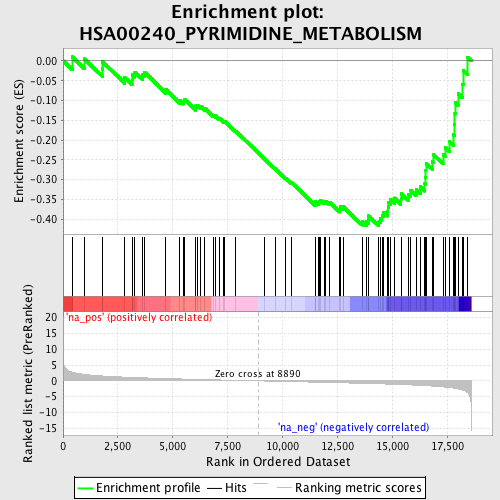
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.41696164 |
| Normalized Enrichment Score (NES) | -1.6335986 |
| Nominal p-value | 0.006722689 |
| FDR q-value | 0.21275303 |
| FWER p-Value | 0.98 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TK1 | 421 | 2.736 | 0.0104 | No | ||
| 2 | NT5C2 | 973 | 2.032 | 0.0052 | No | ||
| 3 | RRM1 | 1788 | 1.537 | -0.0202 | No | ||
| 4 | POLR2J | 1803 | 1.531 | -0.0024 | No | ||
| 5 | POLE | 2803 | 1.182 | -0.0420 | No | ||
| 6 | ENTPD8 | 3144 | 1.087 | -0.0472 | No | ||
| 7 | POLR3A | 3155 | 1.083 | -0.0347 | No | ||
| 8 | NT5E | 3269 | 1.052 | -0.0281 | No | ||
| 9 | UPP2 | 3611 | 0.961 | -0.0348 | No | ||
| 10 | ENTPD6 | 3709 | 0.937 | -0.0287 | No | ||
| 11 | POLA1 | 4685 | 0.719 | -0.0726 | No | ||
| 12 | UPP1 | 5323 | 0.597 | -0.0998 | No | ||
| 13 | POLR2I | 5491 | 0.567 | -0.1019 | No | ||
| 14 | POLR2C | 5512 | 0.562 | -0.0962 | No | ||
| 15 | ENTPD1 | 6036 | 0.467 | -0.1188 | No | ||
| 16 | POLR2B | 6047 | 0.465 | -0.1137 | No | ||
| 17 | ECGF1 | 6120 | 0.452 | -0.1121 | No | ||
| 18 | POLR2E | 6254 | 0.428 | -0.1142 | No | ||
| 19 | DPYD | 6461 | 0.390 | -0.1205 | No | ||
| 20 | TXNRD1 | 6841 | 0.332 | -0.1370 | No | ||
| 21 | TK2 | 6946 | 0.313 | -0.1388 | No | ||
| 22 | POLD4 | 7122 | 0.279 | -0.1449 | No | ||
| 23 | PNPT1 | 7315 | 0.247 | -0.1522 | No | ||
| 24 | POLR2G | 7365 | 0.239 | -0.1520 | No | ||
| 25 | ITPA | 7871 | 0.162 | -0.1773 | No | ||
| 26 | CAD | 9166 | -0.041 | -0.2466 | No | ||
| 27 | POLR1B | 9661 | -0.113 | -0.2718 | No | ||
| 28 | ENTPD4 | 10154 | -0.188 | -0.2961 | No | ||
| 29 | POLE2 | 10390 | -0.222 | -0.3061 | No | ||
| 30 | POLR2K | 11488 | -0.406 | -0.3604 | No | ||
| 31 | DTYMK | 11495 | -0.407 | -0.3558 | No | ||
| 32 | POLR1A | 11641 | -0.431 | -0.3584 | No | ||
| 33 | ENTPD3 | 11666 | -0.435 | -0.3544 | No | ||
| 34 | NT5C | 11738 | -0.446 | -0.3529 | No | ||
| 35 | NME6 | 11890 | -0.468 | -0.3553 | No | ||
| 36 | POLE4 | 11974 | -0.481 | -0.3540 | No | ||
| 37 | DHODH | 12134 | -0.510 | -0.3564 | No | ||
| 38 | NUDT2 | 12603 | -0.597 | -0.3745 | No | ||
| 39 | POLR3H | 12630 | -0.602 | -0.3686 | No | ||
| 40 | CANT1 | 12757 | -0.621 | -0.3679 | No | ||
| 41 | DPYS | 13624 | -0.773 | -0.4052 | No | ||
| 42 | POLD3 | 13814 | -0.807 | -0.4057 | No | ||
| 43 | NT5C3 | 13916 | -0.825 | -0.4012 | No | ||
| 44 | UPB1 | 13923 | -0.827 | -0.3915 | No | ||
| 45 | POLR3K | 14396 | -0.923 | -0.4058 | Yes | ||
| 46 | DCTD | 14462 | -0.936 | -0.3980 | Yes | ||
| 47 | RRM2B | 14535 | -0.953 | -0.3904 | Yes | ||
| 48 | POLE3 | 14606 | -0.969 | -0.3824 | Yes | ||
| 49 | CTPS | 14781 | -1.010 | -0.3796 | Yes | ||
| 50 | PRIM1 | 14807 | -1.018 | -0.3687 | Yes | ||
| 51 | AICDA | 14835 | -1.024 | -0.3578 | Yes | ||
| 52 | POLR2A | 14928 | -1.043 | -0.3501 | Yes | ||
| 53 | POLD2 | 15100 | -1.083 | -0.3463 | Yes | ||
| 54 | POLR2H | 15400 | -1.161 | -0.3484 | Yes | ||
| 55 | NT5C1B | 15413 | -1.165 | -0.3349 | Yes | ||
| 56 | TXNRD2 | 15728 | -1.247 | -0.3368 | Yes | ||
| 57 | ZNRD1 | 15821 | -1.274 | -0.3264 | Yes | ||
| 58 | POLA2 | 16086 | -1.355 | -0.3243 | Yes | ||
| 59 | CTPS2 | 16283 | -1.422 | -0.3176 | Yes | ||
| 60 | NME7 | 16489 | -1.499 | -0.3106 | Yes | ||
| 61 | TYMS | 16506 | -1.506 | -0.2933 | Yes | ||
| 62 | RRM2 | 16534 | -1.516 | -0.2764 | Yes | ||
| 63 | POLR1D | 16540 | -1.517 | -0.2583 | Yes | ||
| 64 | NME4 | 16822 | -1.639 | -0.2537 | Yes | ||
| 65 | POLR3B | 16887 | -1.669 | -0.2370 | Yes | ||
| 66 | CDA | 17317 | -1.906 | -0.2371 | Yes | ||
| 67 | DCK | 17413 | -1.960 | -0.2185 | Yes | ||
| 68 | ENTPD5 | 17596 | -2.106 | -0.2029 | Yes | ||
| 69 | NME2 | 17785 | -2.254 | -0.1858 | Yes | ||
| 70 | NT5M | 17846 | -2.313 | -0.1611 | Yes | ||
| 71 | POLD1 | 17858 | -2.338 | -0.1334 | Yes | ||
| 72 | UMPS | 17871 | -2.352 | -0.1056 | Yes | ||
| 73 | NME1 | 18002 | -2.529 | -0.0821 | Yes | ||
| 74 | RFC5 | 18209 | -2.884 | -0.0584 | Yes | ||
| 75 | UCK1 | 18252 | -2.979 | -0.0246 | Yes | ||
| 76 | NP | 18437 | -3.659 | 0.0097 | Yes |