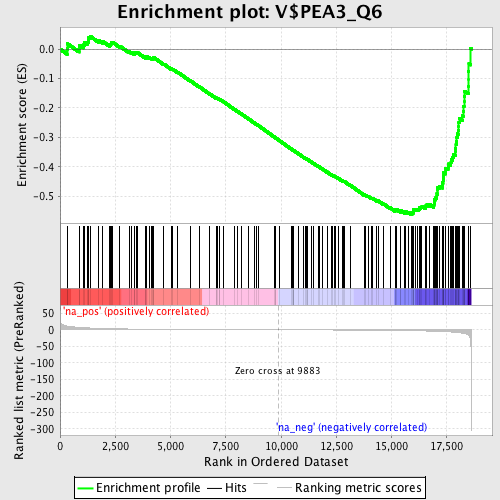
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$PEA3_Q6 |
| Enrichment Score (ES) | -0.56222194 |
| Normalized Enrichment Score (NES) | -1.8088218 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.927 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EXTL2 | 317 | 11.109 | 0.0013 | No | ||
| 2 | SELL | 343 | 10.786 | 0.0179 | No | ||
| 3 | PRDX6 | 857 | 7.187 | 0.0020 | No | ||
| 4 | SLC30A7 | 876 | 7.057 | 0.0128 | No | ||
| 5 | ABLIM1 | 1045 | 6.312 | 0.0142 | No | ||
| 6 | PICALM | 1096 | 6.067 | 0.0216 | No | ||
| 7 | GTF2H1 | 1223 | 5.646 | 0.0241 | No | ||
| 8 | CUGBP1 | 1281 | 5.467 | 0.0301 | No | ||
| 9 | KCNAB2 | 1298 | 5.419 | 0.0383 | No | ||
| 10 | FEN1 | 1352 | 5.246 | 0.0441 | No | ||
| 11 | CENTD2 | 1732 | 4.215 | 0.0306 | No | ||
| 12 | PAFAH2 | 1924 | 3.809 | 0.0266 | No | ||
| 13 | TYSND1 | 2236 | 3.268 | 0.0152 | No | ||
| 14 | SLC2A3 | 2299 | 3.163 | 0.0171 | No | ||
| 15 | DPP3 | 2307 | 3.150 | 0.0219 | No | ||
| 16 | TAF5 | 2366 | 3.050 | 0.0238 | No | ||
| 17 | SLC26A9 | 2702 | 2.659 | 0.0101 | No | ||
| 18 | GPR120 | 3121 | 2.272 | -0.0088 | No | ||
| 19 | GFI1 | 3233 | 2.198 | -0.0111 | No | ||
| 20 | ELAVL4 | 3382 | 2.086 | -0.0157 | No | ||
| 21 | KCNQ1 | 3387 | 2.082 | -0.0124 | No | ||
| 22 | ANK3 | 3462 | 2.033 | -0.0130 | No | ||
| 23 | MMP7 | 3492 | 2.017 | -0.0113 | No | ||
| 24 | TNNI2 | 3860 | 1.804 | -0.0281 | No | ||
| 25 | RPS3 | 3861 | 1.803 | -0.0251 | No | ||
| 26 | DMBT1 | 3912 | 1.776 | -0.0249 | No | ||
| 27 | FGF23 | 4028 | 1.723 | -0.0283 | No | ||
| 28 | LTBR | 4134 | 1.673 | -0.0312 | No | ||
| 29 | TIAL1 | 4197 | 1.643 | -0.0318 | No | ||
| 30 | LZTS2 | 4205 | 1.638 | -0.0295 | No | ||
| 31 | VDAC2 | 4236 | 1.619 | -0.0284 | No | ||
| 32 | BDNF | 4697 | 1.430 | -0.0509 | No | ||
| 33 | NDUFS2 | 5024 | 1.316 | -0.0664 | No | ||
| 34 | CTBP2 | 5089 | 1.291 | -0.0677 | No | ||
| 35 | CYR61 | 5325 | 1.214 | -0.0784 | No | ||
| 36 | MARK1 | 5891 | 1.035 | -0.1073 | No | ||
| 37 | GPA33 | 6297 | 0.912 | -0.1278 | No | ||
| 38 | LAMC1 | 6771 | 0.782 | -0.1521 | No | ||
| 39 | CD3E | 7061 | 0.707 | -0.1666 | No | ||
| 40 | RIN1 | 7072 | 0.706 | -0.1659 | No | ||
| 41 | LAG3 | 7120 | 0.693 | -0.1673 | No | ||
| 42 | RUNX3 | 7201 | 0.672 | -0.1705 | No | ||
| 43 | ANKRD1 | 7213 | 0.670 | -0.1700 | No | ||
| 44 | GATA3 | 7390 | 0.619 | -0.1785 | No | ||
| 45 | CD5 | 7876 | 0.490 | -0.2040 | No | ||
| 46 | LALBA | 8033 | 0.453 | -0.2117 | No | ||
| 47 | VCAM1 | 8195 | 0.411 | -0.2197 | No | ||
| 48 | IL18BP | 8528 | 0.329 | -0.2372 | No | ||
| 49 | PAX6 | 8778 | 0.269 | -0.2502 | No | ||
| 50 | PGM2L1 | 8870 | 0.249 | -0.2547 | No | ||
| 51 | CAPZA1 | 8971 | 0.221 | -0.2598 | No | ||
| 52 | XCL1 | 9724 | 0.044 | -0.3005 | No | ||
| 53 | BCL2L14 | 9739 | 0.040 | -0.3011 | No | ||
| 54 | PRF1 | 9750 | 0.036 | -0.3016 | No | ||
| 55 | GPD1 | 9914 | -0.008 | -0.3104 | No | ||
| 56 | SNCG | 10451 | -0.142 | -0.3392 | No | ||
| 57 | LRFN4 | 10538 | -0.164 | -0.3436 | No | ||
| 58 | FCGR3A | 10560 | -0.170 | -0.3445 | No | ||
| 59 | FUCA1 | 10564 | -0.171 | -0.3444 | No | ||
| 60 | CTSW | 10565 | -0.172 | -0.3441 | No | ||
| 61 | RNASEL | 10772 | -0.226 | -0.3549 | No | ||
| 62 | MMRN2 | 11036 | -0.298 | -0.3686 | No | ||
| 63 | NUMA1 | 11085 | -0.317 | -0.3707 | No | ||
| 64 | EHF | 11135 | -0.327 | -0.3728 | No | ||
| 65 | IPO7 | 11203 | -0.344 | -0.3759 | No | ||
| 66 | SLC41A1 | 11206 | -0.346 | -0.3754 | No | ||
| 67 | HIPK1 | 11395 | -0.398 | -0.3849 | No | ||
| 68 | LIN28 | 11478 | -0.419 | -0.3887 | No | ||
| 69 | SELP | 11697 | -0.481 | -0.3997 | No | ||
| 70 | PLCB3 | 11718 | -0.485 | -0.4000 | No | ||
| 71 | SPRR4 | 11885 | -0.530 | -0.4081 | No | ||
| 72 | RAB39 | 12081 | -0.582 | -0.4177 | No | ||
| 73 | CPNE8 | 12272 | -0.642 | -0.4269 | No | ||
| 74 | NCAM1 | 12337 | -0.661 | -0.4293 | No | ||
| 75 | ADAMTS4 | 12396 | -0.679 | -0.4313 | No | ||
| 76 | SV2A | 12459 | -0.699 | -0.4335 | No | ||
| 77 | GAB2 | 12608 | -0.747 | -0.4402 | No | ||
| 78 | GRIN2B | 12769 | -0.795 | -0.4476 | No | ||
| 79 | SYTL1 | 12840 | -0.822 | -0.4500 | No | ||
| 80 | PDGFD | 12869 | -0.830 | -0.4502 | No | ||
| 81 | MAP4K2 | 13131 | -0.927 | -0.4627 | No | ||
| 82 | CBL | 13764 | -1.164 | -0.4951 | No | ||
| 83 | INPPL1 | 13841 | -1.199 | -0.4972 | No | ||
| 84 | SLC35C1 | 13942 | -1.240 | -0.5005 | No | ||
| 85 | SPATA6 | 14096 | -1.317 | -0.5066 | No | ||
| 86 | TNFRSF19L | 14142 | -1.341 | -0.5068 | No | ||
| 87 | CACNA1E | 14331 | -1.437 | -0.5146 | No | ||
| 88 | IVL | 14397 | -1.471 | -0.5157 | No | ||
| 89 | TRIM33 | 14644 | -1.604 | -0.5264 | No | ||
| 90 | KAZALD1 | 14938 | -1.770 | -0.5393 | No | ||
| 91 | ARF3 | 15160 | -1.919 | -0.5481 | No | ||
| 92 | CTSS | 15176 | -1.929 | -0.5457 | No | ||
| 93 | LCK | 15211 | -1.955 | -0.5443 | No | ||
| 94 | MR1 | 15420 | -2.138 | -0.5520 | No | ||
| 95 | LPXN | 15423 | -2.139 | -0.5486 | No | ||
| 96 | VPS11 | 15603 | -2.302 | -0.5544 | No | ||
| 97 | MFN2 | 15632 | -2.337 | -0.5521 | No | ||
| 98 | ARHGDIB | 15788 | -2.486 | -0.5563 | No | ||
| 99 | SEC24C | 15898 | -2.612 | -0.5579 | Yes | ||
| 100 | DGKZ | 15950 | -2.686 | -0.5562 | Yes | ||
| 101 | TCIRG1 | 15977 | -2.718 | -0.5531 | Yes | ||
| 102 | ELK4 | 15995 | -2.736 | -0.5494 | Yes | ||
| 103 | RPS6KA4 | 15997 | -2.738 | -0.5449 | Yes | ||
| 104 | SRRM1 | 16093 | -2.852 | -0.5453 | Yes | ||
| 105 | ITPR2 | 16186 | -2.985 | -0.5454 | Yes | ||
| 106 | XPNPEP1 | 16243 | -3.087 | -0.5433 | Yes | ||
| 107 | PUM1 | 16271 | -3.135 | -0.5395 | Yes | ||
| 108 | ETV3 | 16323 | -3.216 | -0.5369 | Yes | ||
| 109 | PBXIP1 | 16375 | -3.294 | -0.5342 | Yes | ||
| 110 | ELOVL1 | 16530 | -3.622 | -0.5365 | Yes | ||
| 111 | TMEM24 | 16558 | -3.686 | -0.5319 | Yes | ||
| 112 | FUT11 | 16603 | -3.761 | -0.5280 | Yes | ||
| 113 | ITPKB | 16731 | -4.043 | -0.5282 | Yes | ||
| 114 | CDC14A | 16908 | -4.437 | -0.5303 | Yes | ||
| 115 | ZRANB1 | 16924 | -4.470 | -0.5237 | Yes | ||
| 116 | LRMP | 16940 | -4.496 | -0.5170 | Yes | ||
| 117 | PTPRC | 16945 | -4.513 | -0.5098 | Yes | ||
| 118 | NT5C2 | 17007 | -4.626 | -0.5054 | Yes | ||
| 119 | SIPA1 | 17049 | -4.712 | -0.4998 | Yes | ||
| 120 | ADRBK1 | 17055 | -4.731 | -0.4922 | Yes | ||
| 121 | ZNF289 | 17077 | -4.803 | -0.4853 | Yes | ||
| 122 | BLNK | 17078 | -4.805 | -0.4773 | Yes | ||
| 123 | CD84 | 17094 | -4.865 | -0.4700 | Yes | ||
| 124 | CMAS | 17168 | -5.086 | -0.4655 | Yes | ||
| 125 | IL2RA | 17295 | -5.472 | -0.4633 | Yes | ||
| 126 | MARK2 | 17302 | -5.480 | -0.4545 | Yes | ||
| 127 | IRAK4 | 17330 | -5.552 | -0.4467 | Yes | ||
| 128 | PRKACB | 17345 | -5.621 | -0.4381 | Yes | ||
| 129 | SLAMF9 | 17355 | -5.656 | -0.4292 | Yes | ||
| 130 | ZDHHC5 | 17365 | -5.686 | -0.4203 | Yes | ||
| 131 | CD69 | 17448 | -5.917 | -0.4149 | Yes | ||
| 132 | AKT3 | 17454 | -5.931 | -0.4053 | Yes | ||
| 133 | POU2AF1 | 17559 | -6.274 | -0.4005 | Yes | ||
| 134 | UNC93B1 | 17569 | -6.302 | -0.3905 | Yes | ||
| 135 | ST7L | 17690 | -6.710 | -0.3858 | Yes | ||
| 136 | PIK4CB | 17701 | -6.755 | -0.3751 | Yes | ||
| 137 | SORL1 | 17771 | -7.017 | -0.3672 | Yes | ||
| 138 | SPI1 | 17810 | -7.163 | -0.3574 | Yes | ||
| 139 | ESRRA | 17890 | -7.595 | -0.3490 | Yes | ||
| 140 | RHOG | 17897 | -7.651 | -0.3366 | Yes | ||
| 141 | TRIM8 | 17917 | -7.741 | -0.3248 | Yes | ||
| 142 | MAPKAPK2 | 17938 | -7.889 | -0.3127 | Yes | ||
| 143 | SSBP3 | 17957 | -7.973 | -0.3005 | Yes | ||
| 144 | TCEB3 | 17984 | -8.205 | -0.2882 | Yes | ||
| 145 | CAMK1D | 18016 | -8.393 | -0.2760 | Yes | ||
| 146 | CD53 | 18040 | -8.507 | -0.2631 | Yes | ||
| 147 | CDKN2C | 18047 | -8.548 | -0.2492 | Yes | ||
| 148 | DCLRE1C | 18067 | -8.658 | -0.2358 | Yes | ||
| 149 | EPHA2 | 18212 | -10.078 | -0.2268 | Yes | ||
| 150 | NCF2 | 18244 | -10.357 | -0.2113 | Yes | ||
| 151 | EPC1 | 18267 | -10.496 | -0.1950 | Yes | ||
| 152 | FLI1 | 18285 | -10.619 | -0.1783 | Yes | ||
| 153 | HHEX | 18291 | -10.672 | -0.1608 | Yes | ||
| 154 | POLD4 | 18311 | -10.982 | -0.1436 | Yes | ||
| 155 | WASF2 | 18473 | -15.094 | -0.1272 | Yes | ||
| 156 | NRGN | 18481 | -15.334 | -0.1021 | Yes | ||
| 157 | CAP1 | 18484 | -15.486 | -0.0765 | Yes | ||
| 158 | PTPN6 | 18504 | -16.488 | -0.0501 | Yes | ||
| 159 | IL18 | 18579 | -33.750 | 0.0020 | Yes |