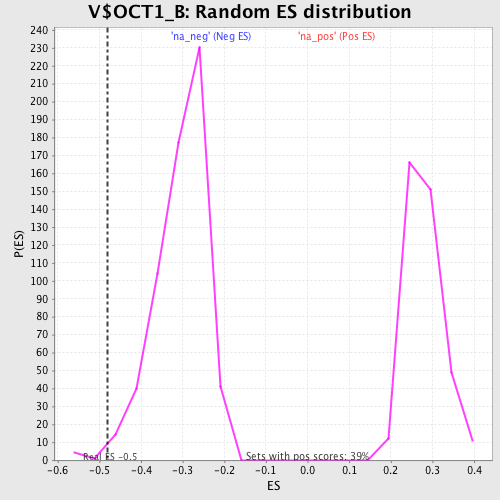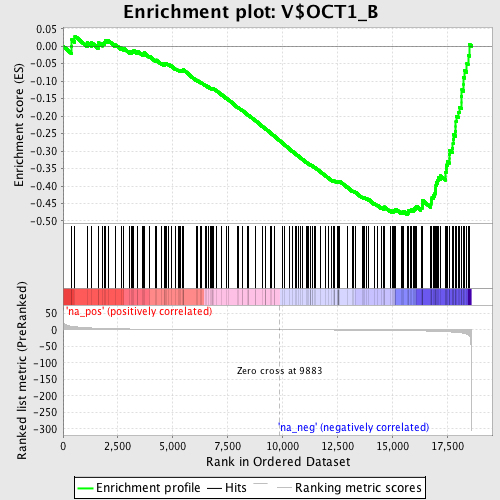
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$OCT1_B |
| Enrichment Score (ES) | -0.4814512 |
| Normalized Enrichment Score (NES) | -1.5873315 |
| Nominal p-value | 0.009819968 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NR1D1 | 387 | 10.397 | -0.0008 | No | ||
| 2 | SERTAD4 | 398 | 10.328 | 0.0186 | No | ||
| 3 | NEDD4 | 538 | 9.224 | 0.0290 | No | ||
| 4 | KCNN3 | 1102 | 6.045 | 0.0101 | No | ||
| 5 | PPARGC1B | 1288 | 5.449 | 0.0107 | No | ||
| 6 | NRXN1 | 1598 | 4.517 | 0.0027 | No | ||
| 7 | LDB2 | 1628 | 4.433 | 0.0097 | No | ||
| 8 | CDK2 | 1797 | 4.060 | 0.0084 | No | ||
| 9 | CD86 | 1882 | 3.880 | 0.0114 | No | ||
| 10 | TMOD2 | 1921 | 3.811 | 0.0167 | No | ||
| 11 | BCL2 | 2046 | 3.584 | 0.0169 | No | ||
| 12 | TBR1 | 2396 | 3.021 | 0.0039 | No | ||
| 13 | E2F3 | 2656 | 2.722 | -0.0049 | No | ||
| 14 | SFRP2 | 2772 | 2.583 | -0.0062 | No | ||
| 15 | NEUROD6 | 3042 | 2.352 | -0.0162 | No | ||
| 16 | CDH13 | 3114 | 2.278 | -0.0156 | No | ||
| 17 | OLIG3 | 3172 | 2.230 | -0.0144 | No | ||
| 18 | BCDO2 | 3224 | 2.201 | -0.0129 | No | ||
| 19 | SCOC | 3369 | 2.092 | -0.0167 | No | ||
| 20 | HOXA10 | 3398 | 2.074 | -0.0142 | No | ||
| 21 | TSGA14 | 3625 | 1.927 | -0.0227 | No | ||
| 22 | MID1 | 3678 | 1.896 | -0.0218 | No | ||
| 23 | NR6A1 | 3687 | 1.892 | -0.0186 | No | ||
| 24 | MGAT5B | 3946 | 1.764 | -0.0292 | No | ||
| 25 | HOXA13 | 4220 | 1.626 | -0.0408 | No | ||
| 26 | LRCH4 | 4251 | 1.610 | -0.0393 | No | ||
| 27 | TWIST1 | 4489 | 1.514 | -0.0493 | No | ||
| 28 | PTHLH | 4608 | 1.464 | -0.0528 | No | ||
| 29 | SOX2 | 4628 | 1.459 | -0.0510 | No | ||
| 30 | HIST2H2AC | 4655 | 1.449 | -0.0496 | No | ||
| 31 | BDNF | 4697 | 1.430 | -0.0491 | No | ||
| 32 | FBXO24 | 4787 | 1.399 | -0.0512 | No | ||
| 33 | TCF2 | 4928 | 1.344 | -0.0562 | No | ||
| 34 | WNT6 | 5139 | 1.274 | -0.0651 | No | ||
| 35 | SLC19A3 | 5238 | 1.244 | -0.0680 | No | ||
| 36 | CYR61 | 5325 | 1.214 | -0.0703 | No | ||
| 37 | IRX4 | 5349 | 1.203 | -0.0693 | No | ||
| 38 | KLF12 | 5423 | 1.180 | -0.0709 | No | ||
| 39 | SLC1A2 | 5465 | 1.170 | -0.0709 | No | ||
| 40 | COL25A1 | 5466 | 1.170 | -0.0686 | No | ||
| 41 | VSNL1 | 5472 | 1.169 | -0.0666 | No | ||
| 42 | HOXB6 | 6060 | 0.987 | -0.0966 | No | ||
| 43 | NRL | 6144 | 0.961 | -0.0992 | No | ||
| 44 | CDX4 | 6256 | 0.924 | -0.1034 | No | ||
| 45 | SP6 | 6326 | 0.903 | -0.1054 | No | ||
| 46 | DNAH5 | 6466 | 0.863 | -0.1113 | No | ||
| 47 | LIPG | 6516 | 0.849 | -0.1123 | No | ||
| 48 | POU3F4 | 6615 | 0.820 | -0.1160 | No | ||
| 49 | LRRTM4 | 6722 | 0.793 | -0.1203 | No | ||
| 50 | SILV | 6768 | 0.782 | -0.1212 | No | ||
| 51 | LHX6 | 6795 | 0.775 | -0.1211 | No | ||
| 52 | RRM2B | 6818 | 0.770 | -0.1208 | No | ||
| 53 | NFIA | 6866 | 0.757 | -0.1219 | No | ||
| 54 | PRDM1 | 6981 | 0.728 | -0.1266 | No | ||
| 55 | IFIT2 | 7225 | 0.666 | -0.1385 | No | ||
| 56 | CDX2 | 7424 | 0.610 | -0.1481 | No | ||
| 57 | RGL1 | 7538 | 0.578 | -0.1531 | No | ||
| 58 | PRKG1 | 7951 | 0.472 | -0.1745 | No | ||
| 59 | LPL | 7987 | 0.464 | -0.1755 | No | ||
| 60 | PCDH20 | 8155 | 0.420 | -0.1838 | No | ||
| 61 | EPHB3 | 8162 | 0.418 | -0.1833 | No | ||
| 62 | ITSN1 | 8411 | 0.359 | -0.1961 | No | ||
| 63 | ALDH1A1 | 8445 | 0.349 | -0.1972 | No | ||
| 64 | FSTL5 | 8471 | 0.343 | -0.1979 | No | ||
| 65 | PCYT1B | 8764 | 0.275 | -0.2132 | No | ||
| 66 | PAX6 | 8778 | 0.269 | -0.2134 | No | ||
| 67 | UBE2S | 9073 | 0.200 | -0.2289 | No | ||
| 68 | NRAS | 9203 | 0.170 | -0.2356 | No | ||
| 69 | TBXAS1 | 9208 | 0.169 | -0.2355 | No | ||
| 70 | ALK | 9244 | 0.160 | -0.2371 | No | ||
| 71 | ADORA2A | 9456 | 0.101 | -0.2483 | No | ||
| 72 | GNB3 | 9487 | 0.096 | -0.2498 | No | ||
| 73 | NXPH1 | 9637 | 0.064 | -0.2577 | No | ||
| 74 | UPK3A | 9642 | 0.063 | -0.2578 | No | ||
| 75 | TSC1 | 9979 | -0.026 | -0.2760 | No | ||
| 76 | TCEAL1 | 10101 | -0.057 | -0.2825 | No | ||
| 77 | SYNPR | 10295 | -0.099 | -0.2927 | No | ||
| 78 | GUCA1A | 10458 | -0.145 | -0.3013 | No | ||
| 79 | GPRC5B | 10600 | -0.178 | -0.3086 | No | ||
| 80 | HPCAL4 | 10635 | -0.189 | -0.3100 | No | ||
| 81 | LYN | 10706 | -0.211 | -0.3134 | No | ||
| 82 | KIF13A | 10807 | -0.236 | -0.3184 | No | ||
| 83 | CNNM4 | 10911 | -0.263 | -0.3235 | No | ||
| 84 | MRAS | 11106 | -0.321 | -0.3334 | No | ||
| 85 | EHF | 11135 | -0.327 | -0.3343 | No | ||
| 86 | RGS13 | 11170 | -0.337 | -0.3355 | No | ||
| 87 | THRA | 11254 | -0.362 | -0.3393 | No | ||
| 88 | SH3GL3 | 11268 | -0.366 | -0.3393 | No | ||
| 89 | KCTD6 | 11291 | -0.371 | -0.3397 | No | ||
| 90 | SKIL | 11346 | -0.383 | -0.3419 | No | ||
| 91 | SLC24A1 | 11375 | -0.389 | -0.3427 | No | ||
| 92 | STAG1 | 11462 | -0.415 | -0.3465 | No | ||
| 93 | HIST1H2AC | 11520 | -0.431 | -0.3488 | No | ||
| 94 | CSF3 | 11725 | -0.488 | -0.3589 | No | ||
| 95 | STAT4 | 11939 | -0.543 | -0.3694 | No | ||
| 96 | FZD2 | 12086 | -0.583 | -0.3762 | No | ||
| 97 | SRF | 12241 | -0.633 | -0.3834 | No | ||
| 98 | GPC4 | 12303 | -0.650 | -0.3854 | No | ||
| 99 | NEUROG1 | 12351 | -0.666 | -0.3867 | No | ||
| 100 | PRRX1 | 12355 | -0.667 | -0.3855 | No | ||
| 101 | MITF | 12381 | -0.676 | -0.3856 | No | ||
| 102 | FGF14 | 12487 | -0.707 | -0.3899 | No | ||
| 103 | PIPOX | 12506 | -0.714 | -0.3895 | No | ||
| 104 | CLCA3 | 12540 | -0.725 | -0.3899 | No | ||
| 105 | POU2F3 | 12551 | -0.728 | -0.3890 | No | ||
| 106 | HOXC5 | 12562 | -0.733 | -0.3881 | No | ||
| 107 | SATB2 | 12568 | -0.734 | -0.3870 | No | ||
| 108 | SLITRK6 | 12573 | -0.735 | -0.3858 | No | ||
| 109 | GAB2 | 12608 | -0.747 | -0.3862 | No | ||
| 110 | DMD | 12944 | -0.850 | -0.4027 | No | ||
| 111 | CSMD3 | 13191 | -0.945 | -0.4142 | No | ||
| 112 | BAT2 | 13228 | -0.958 | -0.4143 | No | ||
| 113 | DPYSL3 | 13318 | -0.988 | -0.4172 | No | ||
| 114 | ASCL3 | 13628 | -1.115 | -0.4318 | No | ||
| 115 | SPRR2B | 13696 | -1.131 | -0.4333 | No | ||
| 116 | SREBF2 | 13743 | -1.155 | -0.4335 | No | ||
| 117 | FGF12 | 13830 | -1.192 | -0.4359 | No | ||
| 118 | GPR4 | 13917 | -1.226 | -0.4382 | No | ||
| 119 | SESN3 | 14187 | -1.360 | -0.4501 | No | ||
| 120 | PFKFB1 | 14318 | -1.431 | -0.4544 | No | ||
| 121 | LRRN1 | 14521 | -1.540 | -0.4624 | No | ||
| 122 | OTX2 | 14612 | -1.587 | -0.4642 | No | ||
| 123 | ITPR3 | 14626 | -1.593 | -0.4618 | No | ||
| 124 | NRXN3 | 14648 | -1.606 | -0.4598 | No | ||
| 125 | HIST1H3C | 14907 | -1.749 | -0.4704 | No | ||
| 126 | TRPM1 | 15032 | -1.842 | -0.4736 | No | ||
| 127 | RAB26 | 15054 | -1.857 | -0.4711 | No | ||
| 128 | CRYZL1 | 15111 | -1.888 | -0.4705 | No | ||
| 129 | SLC6A15 | 15146 | -1.910 | -0.4687 | No | ||
| 130 | CRISP1 | 15162 | -1.921 | -0.4658 | No | ||
| 131 | ADORA1 | 15417 | -2.136 | -0.4754 | No | ||
| 132 | CPD | 15475 | -2.179 | -0.4743 | No | ||
| 133 | KCTD12 | 15522 | -2.230 | -0.4724 | No | ||
| 134 | COL23A1 | 15689 | -2.387 | -0.4768 | Yes | ||
| 135 | NDNL2 | 15725 | -2.432 | -0.4740 | Yes | ||
| 136 | PDE4D | 15754 | -2.452 | -0.4708 | Yes | ||
| 137 | TLL2 | 15815 | -2.516 | -0.4692 | Yes | ||
| 138 | HIST1H2AH | 15886 | -2.595 | -0.4679 | Yes | ||
| 139 | PIM2 | 15964 | -2.697 | -0.4669 | Yes | ||
| 140 | RHOB | 16003 | -2.745 | -0.4636 | Yes | ||
| 141 | HIST1H2AB | 16072 | -2.832 | -0.4618 | Yes | ||
| 142 | EAF2 | 16095 | -2.855 | -0.4575 | Yes | ||
| 143 | H3F3B | 16311 | -3.201 | -0.4629 | Yes | ||
| 144 | REL | 16362 | -3.269 | -0.4593 | Yes | ||
| 145 | SH3BGRL | 16366 | -3.277 | -0.4531 | Yes | ||
| 146 | AXUD1 | 16377 | -3.295 | -0.4473 | Yes | ||
| 147 | GPM6A | 16380 | -3.304 | -0.4410 | Yes | ||
| 148 | POLM | 16762 | -4.123 | -0.4537 | Yes | ||
| 149 | HIST1H2BC | 16769 | -4.137 | -0.4460 | Yes | ||
| 150 | NR2C2 | 16775 | -4.148 | -0.4382 | Yes | ||
| 151 | PIK3CD | 16811 | -4.218 | -0.4319 | Yes | ||
| 152 | ZHX2 | 16890 | -4.381 | -0.4277 | Yes | ||
| 153 | MBIP | 16943 | -4.511 | -0.4217 | Yes | ||
| 154 | CSNK1E | 16965 | -4.536 | -0.4141 | Yes | ||
| 155 | BIN3 | 16982 | -4.575 | -0.4061 | Yes | ||
| 156 | MAP2K6 | 16991 | -4.589 | -0.3976 | Yes | ||
| 157 | DLGAP4 | 17018 | -4.646 | -0.3900 | Yes | ||
| 158 | BLNK | 17078 | -4.805 | -0.3839 | Yes | ||
| 159 | BTK | 17106 | -4.910 | -0.3759 | Yes | ||
| 160 | HDAC9 | 17200 | -5.177 | -0.3709 | Yes | ||
| 161 | PTEN | 17426 | -5.855 | -0.3717 | Yes | ||
| 162 | HIST1H3B | 17427 | -5.857 | -0.3604 | Yes | ||
| 163 | NOTCH1 | 17461 | -5.950 | -0.3506 | Yes | ||
| 164 | HIST1H2BK | 17476 | -5.969 | -0.3398 | Yes | ||
| 165 | FOXP1 | 17514 | -6.087 | -0.3300 | Yes | ||
| 166 | ARPC5 | 17596 | -6.399 | -0.3220 | Yes | ||
| 167 | NFYB | 17598 | -6.408 | -0.3097 | Yes | ||
| 168 | HIST2H2BE | 17623 | -6.502 | -0.2984 | Yes | ||
| 169 | GFI1B | 17750 | -6.918 | -0.2918 | Yes | ||
| 170 | SLC25A12 | 17769 | -7.009 | -0.2792 | Yes | ||
| 171 | THRAP5 | 17779 | -7.069 | -0.2660 | Yes | ||
| 172 | FES | 17794 | -7.117 | -0.2529 | Yes | ||
| 173 | ESRRA | 17890 | -7.595 | -0.2434 | Yes | ||
| 174 | ARHGAP4 | 17902 | -7.667 | -0.2291 | Yes | ||
| 175 | HIST1H2BB | 17904 | -7.678 | -0.2143 | Yes | ||
| 176 | HIST1H2BM | 17940 | -7.891 | -0.2009 | Yes | ||
| 177 | RHOBTB2 | 18022 | -8.436 | -0.1889 | Yes | ||
| 178 | ETV1 | 18059 | -8.622 | -0.1742 | Yes | ||
| 179 | DUSP6 | 18160 | -9.525 | -0.1611 | Yes | ||
| 180 | TCF12 | 18161 | -9.536 | -0.1426 | Yes | ||
| 181 | CD79B | 18176 | -9.666 | -0.1247 | Yes | ||
| 182 | SCML4 | 18252 | -10.384 | -0.1086 | Yes | ||
| 183 | ATF7IP | 18257 | -10.399 | -0.0887 | Yes | ||
| 184 | RRAS | 18306 | -10.843 | -0.0702 | Yes | ||
| 185 | VPREB3 | 18402 | -12.838 | -0.0505 | Yes | ||
| 186 | EGR2 | 18458 | -14.584 | -0.0252 | Yes | ||
| 187 | ACBD4 | 18515 | -17.407 | 0.0055 | Yes |