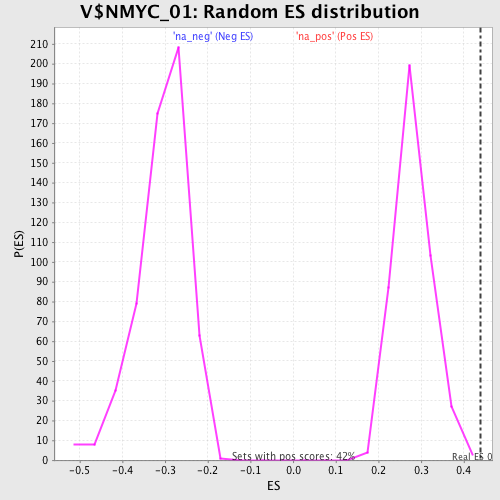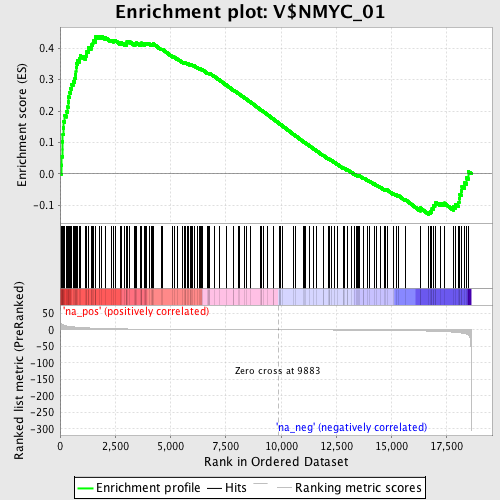
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$NMYC_01 |
| Enrichment Score (ES) | 0.4386861 |
| Normalized Enrichment Score (NES) | 1.5592152 |
| Nominal p-value | 0.0023640662 |
| FDR q-value | 0.047477268 |
| FWER p-Value | 0.84 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RCL1 | 46 | 19.200 | 0.0284 | Yes | ||
| 2 | FBL | 68 | 17.727 | 0.0559 | Yes | ||
| 3 | POLR3E | 109 | 15.120 | 0.0781 | Yes | ||
| 4 | PPAT | 110 | 15.096 | 0.1024 | Yes | ||
| 5 | IPO4 | 127 | 14.637 | 0.1251 | Yes | ||
| 6 | ATAD3A | 148 | 14.035 | 0.1466 | Yes | ||
| 7 | G6PC3 | 166 | 13.392 | 0.1673 | Yes | ||
| 8 | HSPD1 | 213 | 12.457 | 0.1849 | Yes | ||
| 9 | ATP5G1 | 284 | 11.503 | 0.1996 | Yes | ||
| 10 | HRSP12 | 350 | 10.707 | 0.2133 | Yes | ||
| 11 | LIG3 | 366 | 10.549 | 0.2295 | Yes | ||
| 12 | NR1D1 | 387 | 10.397 | 0.2452 | Yes | ||
| 13 | COPS7A | 437 | 9.932 | 0.2585 | Yes | ||
| 14 | TIMM10 | 474 | 9.631 | 0.2721 | Yes | ||
| 15 | PABPC4 | 519 | 9.355 | 0.2848 | Yes | ||
| 16 | MTHFD1 | 589 | 8.792 | 0.2952 | Yes | ||
| 17 | PA2G4 | 666 | 8.313 | 0.3045 | Yes | ||
| 18 | XRN2 | 700 | 8.032 | 0.3156 | Yes | ||
| 19 | UBE1 | 718 | 7.942 | 0.3275 | Yes | ||
| 20 | VARS2 | 720 | 7.927 | 0.3402 | Yes | ||
| 21 | HSPE1 | 735 | 7.831 | 0.3521 | Yes | ||
| 22 | AMPD2 | 789 | 7.536 | 0.3613 | Yes | ||
| 23 | FKBP5 | 889 | 6.987 | 0.3672 | Yes | ||
| 24 | ZZZ3 | 907 | 6.928 | 0.3775 | Yes | ||
| 25 | GPS1 | 1136 | 5.929 | 0.3746 | Yes | ||
| 26 | PRDX4 | 1180 | 5.811 | 0.3817 | Yes | ||
| 27 | NOL5A | 1208 | 5.699 | 0.3894 | Yes | ||
| 28 | CUGBP1 | 1281 | 5.467 | 0.3943 | Yes | ||
| 29 | PPARGC1B | 1288 | 5.449 | 0.4027 | Yes | ||
| 30 | HYAL2 | 1402 | 5.051 | 0.4048 | Yes | ||
| 31 | RXRB | 1437 | 4.949 | 0.4109 | Yes | ||
| 32 | RAMP2 | 1485 | 4.790 | 0.4161 | Yes | ||
| 33 | PANK3 | 1490 | 4.774 | 0.4235 | Yes | ||
| 34 | PABPC1 | 1592 | 4.530 | 0.4254 | Yes | ||
| 35 | NRXN1 | 1598 | 4.517 | 0.4324 | Yes | ||
| 36 | EIF4G1 | 1615 | 4.467 | 0.4387 | Yes | ||
| 37 | MAPKAPK3 | 1766 | 4.135 | 0.4372 | No | ||
| 38 | SFXN2 | 1872 | 3.898 | 0.4378 | No | ||
| 39 | BCL2 | 2046 | 3.584 | 0.4342 | No | ||
| 40 | ELOVL6 | 2308 | 3.149 | 0.4251 | No | ||
| 41 | EIF3S1 | 2409 | 3.006 | 0.4245 | No | ||
| 42 | HTF9C | 2494 | 2.917 | 0.4247 | No | ||
| 43 | SLC20A1 | 2720 | 2.638 | 0.4167 | No | ||
| 44 | SOX12 | 2785 | 2.571 | 0.4174 | No | ||
| 45 | BCL7C | 2929 | 2.453 | 0.4136 | No | ||
| 46 | HMOX1 | 3016 | 2.371 | 0.4127 | No | ||
| 47 | SRP72 | 3020 | 2.369 | 0.4164 | No | ||
| 48 | EIF4B | 3024 | 2.367 | 0.4201 | No | ||
| 49 | NEUROD6 | 3042 | 2.352 | 0.4229 | No | ||
| 50 | WEE1 | 3159 | 2.243 | 0.4202 | No | ||
| 51 | TIMM9 | 3377 | 2.087 | 0.4118 | No | ||
| 52 | NR5A1 | 3407 | 2.064 | 0.4136 | No | ||
| 53 | NPM1 | 3426 | 2.056 | 0.4159 | No | ||
| 54 | STMN4 | 3463 | 2.032 | 0.4172 | No | ||
| 55 | CHST11 | 3626 | 1.926 | 0.4116 | No | ||
| 56 | ADCY3 | 3667 | 1.902 | 0.4125 | No | ||
| 57 | RTN4RL2 | 3694 | 1.890 | 0.4141 | No | ||
| 58 | U2AF2 | 3699 | 1.888 | 0.4169 | No | ||
| 59 | EN2 | 3830 | 1.820 | 0.4128 | No | ||
| 60 | ANGPT2 | 3871 | 1.796 | 0.4135 | No | ||
| 61 | CEBPB | 3882 | 1.792 | 0.4159 | No | ||
| 62 | PLA2G4A | 3922 | 1.774 | 0.4166 | No | ||
| 63 | HRH3 | 4025 | 1.725 | 0.4139 | No | ||
| 64 | LHX9 | 4119 | 1.683 | 0.4115 | No | ||
| 65 | FLT3 | 4175 | 1.654 | 0.4112 | No | ||
| 66 | TIAL1 | 4197 | 1.643 | 0.4127 | No | ||
| 67 | PES1 | 4217 | 1.629 | 0.4143 | No | ||
| 68 | POGK | 4576 | 1.479 | 0.3973 | No | ||
| 69 | BARHL1 | 4618 | 1.462 | 0.3974 | No | ||
| 70 | CTBP2 | 5089 | 1.291 | 0.3740 | No | ||
| 71 | KRTCAP2 | 5160 | 1.268 | 0.3723 | No | ||
| 72 | KCNK5 | 5312 | 1.218 | 0.3660 | No | ||
| 73 | RPL13A | 5550 | 1.142 | 0.3550 | No | ||
| 74 | CLCN2 | 5630 | 1.116 | 0.3525 | No | ||
| 75 | RBM3 | 5666 | 1.103 | 0.3524 | No | ||
| 76 | KIAA0427 | 5680 | 1.098 | 0.3535 | No | ||
| 77 | SGTB | 5753 | 1.076 | 0.3513 | No | ||
| 78 | NDUFS1 | 5802 | 1.062 | 0.3504 | No | ||
| 79 | IPO13 | 5879 | 1.039 | 0.3480 | No | ||
| 80 | ADAMTS17 | 5901 | 1.033 | 0.3485 | No | ||
| 81 | ALX4 | 5966 | 1.017 | 0.3467 | No | ||
| 82 | PRKCE | 6003 | 1.007 | 0.3463 | No | ||
| 83 | ADPRHL1 | 6063 | 0.985 | 0.3447 | No | ||
| 84 | RUNX2 | 6201 | 0.942 | 0.3388 | No | ||
| 85 | SEMA7A | 6291 | 0.914 | 0.3354 | No | ||
| 86 | DLC1 | 6333 | 0.902 | 0.3347 | No | ||
| 87 | TPM2 | 6406 | 0.880 | 0.3322 | No | ||
| 88 | B3GALT2 | 6453 | 0.866 | 0.3311 | No | ||
| 89 | SNCAIP | 6672 | 0.806 | 0.3206 | No | ||
| 90 | HOXC13 | 6713 | 0.794 | 0.3197 | No | ||
| 91 | CACNA1D | 6718 | 0.794 | 0.3207 | No | ||
| 92 | LMNB1 | 6772 | 0.781 | 0.3191 | No | ||
| 93 | ATP6V0B | 6777 | 0.781 | 0.3202 | No | ||
| 94 | FGF11 | 6969 | 0.732 | 0.3110 | No | ||
| 95 | ENPP6 | 7197 | 0.674 | 0.2998 | No | ||
| 96 | CTSF | 7509 | 0.586 | 0.2838 | No | ||
| 97 | SLC12A5 | 7839 | 0.499 | 0.2668 | No | ||
| 98 | PCDHA10 | 7868 | 0.492 | 0.2661 | No | ||
| 99 | ZCCHC7 | 8077 | 0.438 | 0.2555 | No | ||
| 100 | SEPT3 | 8127 | 0.426 | 0.2535 | No | ||
| 101 | RBBP6 | 8335 | 0.379 | 0.2429 | No | ||
| 102 | GPC3 | 8430 | 0.353 | 0.2384 | No | ||
| 103 | NKX2-2 | 8634 | 0.305 | 0.2279 | No | ||
| 104 | NEUROD2 | 9085 | 0.197 | 0.2038 | No | ||
| 105 | NUP153 | 9114 | 0.191 | 0.2026 | No | ||
| 106 | TESK2 | 9115 | 0.191 | 0.2029 | No | ||
| 107 | NRAS | 9203 | 0.170 | 0.1984 | No | ||
| 108 | DLX1 | 9366 | 0.128 | 0.1898 | No | ||
| 109 | NXPH1 | 9637 | 0.064 | 0.1753 | No | ||
| 110 | MNT | 9650 | 0.062 | 0.1748 | No | ||
| 111 | GPD1 | 9914 | -0.008 | 0.1605 | No | ||
| 112 | CHRM1 | 9946 | -0.018 | 0.1589 | No | ||
| 113 | MYST3 | 9987 | -0.028 | 0.1567 | No | ||
| 114 | SCFD2 | 9988 | -0.029 | 0.1568 | No | ||
| 115 | PLAGL1 | 10087 | -0.053 | 0.1515 | No | ||
| 116 | APOA5 | 10575 | -0.174 | 0.1254 | No | ||
| 117 | JPH1 | 10669 | -0.200 | 0.1207 | No | ||
| 118 | COL2A1 | 11018 | -0.293 | 0.1023 | No | ||
| 119 | HMGN2 | 11070 | -0.312 | 0.1000 | No | ||
| 120 | HPCA | 11111 | -0.323 | 0.0984 | No | ||
| 121 | ADAM10 | 11302 | -0.374 | 0.0887 | No | ||
| 122 | LIN28 | 11478 | -0.419 | 0.0799 | No | ||
| 123 | HOXB7 | 11617 | -0.461 | 0.0731 | No | ||
| 124 | HHIP | 11900 | -0.536 | 0.0587 | No | ||
| 125 | POU3F1 | 11903 | -0.537 | 0.0594 | No | ||
| 126 | ABCA1 | 12124 | -0.596 | 0.0485 | No | ||
| 127 | DNAJB9 | 12176 | -0.614 | 0.0467 | No | ||
| 128 | MCM8 | 12180 | -0.614 | 0.0475 | No | ||
| 129 | SC5DL | 12279 | -0.643 | 0.0433 | No | ||
| 130 | RPL22 | 12421 | -0.687 | 0.0367 | No | ||
| 131 | HS3ST3A1 | 12554 | -0.729 | 0.0307 | No | ||
| 132 | RNF128 | 12808 | -0.810 | 0.0183 | No | ||
| 133 | PFN1 | 12820 | -0.816 | 0.0190 | No | ||
| 134 | RPIA | 12876 | -0.832 | 0.0174 | No | ||
| 135 | GRK6 | 12986 | -0.867 | 0.0129 | No | ||
| 136 | SEMA3F | 13029 | -0.886 | 0.0120 | No | ||
| 137 | SLC38A5 | 13186 | -0.943 | 0.0051 | No | ||
| 138 | LAP3 | 13317 | -0.988 | -0.0004 | No | ||
| 139 | B4GALT2 | 13418 | -1.023 | -0.0042 | No | ||
| 140 | TLE3 | 13477 | -1.055 | -0.0056 | No | ||
| 141 | DNMT3A | 13484 | -1.060 | -0.0042 | No | ||
| 142 | VPS16 | 13542 | -1.083 | -0.0056 | No | ||
| 143 | SLC17A2 | 13724 | -1.144 | -0.0135 | No | ||
| 144 | USP2 | 13751 | -1.160 | -0.0131 | No | ||
| 145 | PLAG1 | 13919 | -1.229 | -0.0202 | No | ||
| 146 | TOP1 | 14010 | -1.275 | -0.0230 | No | ||
| 147 | BAI2 | 14208 | -1.372 | -0.0315 | No | ||
| 148 | DUSP4 | 14333 | -1.439 | -0.0359 | No | ||
| 149 | FKBP10 | 14494 | -1.526 | -0.0421 | No | ||
| 150 | FHOD1 | 14694 | -1.634 | -0.0502 | No | ||
| 151 | PDK2 | 14720 | -1.649 | -0.0489 | No | ||
| 152 | GRIN2A | 14807 | -1.691 | -0.0509 | No | ||
| 153 | ARF6 | 15099 | -1.881 | -0.0636 | No | ||
| 154 | GTPBP1 | 15209 | -1.954 | -0.0664 | No | ||
| 155 | MAX | 15307 | -2.029 | -0.0684 | No | ||
| 156 | ANXA6 | 15625 | -2.330 | -0.0818 | No | ||
| 157 | SCYL1 | 16296 | -3.176 | -0.1130 | No | ||
| 158 | AKAP10 | 16299 | -3.183 | -0.1080 | No | ||
| 159 | GADD45G | 16689 | -3.925 | -0.1228 | No | ||
| 160 | PFKFB3 | 16773 | -4.143 | -0.1206 | No | ||
| 161 | BMP2K | 16823 | -4.242 | -0.1164 | No | ||
| 162 | TEF | 16830 | -4.261 | -0.1099 | No | ||
| 163 | ZHX2 | 16890 | -4.381 | -0.1060 | No | ||
| 164 | CDC14A | 16908 | -4.437 | -0.0998 | No | ||
| 165 | MXD4 | 16968 | -4.544 | -0.0957 | No | ||
| 166 | RAD9A | 17005 | -4.624 | -0.0902 | No | ||
| 167 | GADD45B | 17216 | -5.240 | -0.0931 | No | ||
| 168 | GNA13 | 17375 | -5.722 | -0.0925 | No | ||
| 169 | ARRDC3 | 17795 | -7.118 | -0.1037 | No | ||
| 170 | PSEN2 | 17891 | -7.601 | -0.0967 | No | ||
| 171 | SH3KBP1 | 18046 | -8.546 | -0.0912 | No | ||
| 172 | ETV1 | 18059 | -8.622 | -0.0780 | No | ||
| 173 | SOCS5 | 18091 | -8.885 | -0.0654 | No | ||
| 174 | WBP2 | 18159 | -9.500 | -0.0537 | No | ||
| 175 | STAT3 | 18184 | -9.748 | -0.0393 | No | ||
| 176 | AP1GBP1 | 18313 | -10.997 | -0.0285 | No | ||
| 177 | GJA1 | 18398 | -12.744 | -0.0125 | No | ||
| 178 | HBP1 | 18472 | -15.082 | 0.0078 | No |