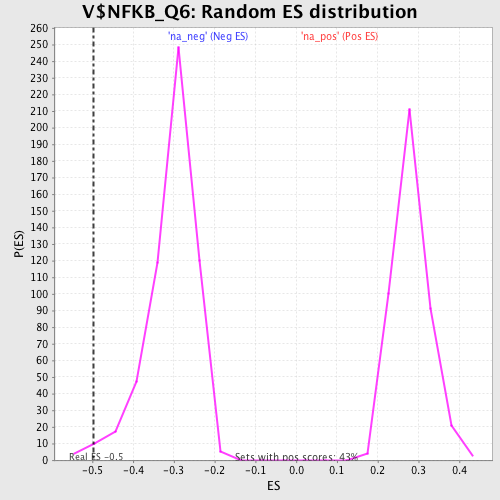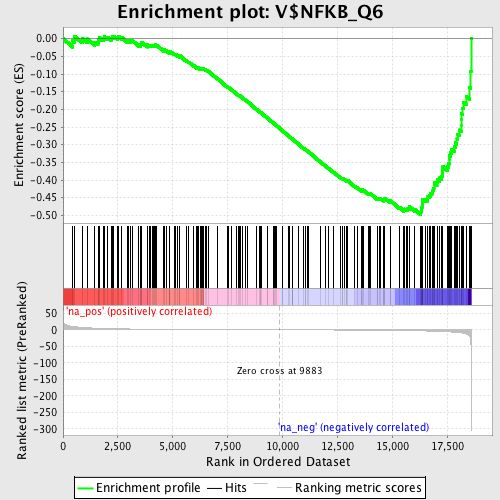
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFKB_Q6 |
| Enrichment Score (ES) | -0.4974701 |
| Normalized Enrichment Score (NES) | -1.6303722 |
| Nominal p-value | 0.015789473 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DAP3 | 441 | 9.881 | -0.0058 | No | ||
| 2 | PCBP4 | 534 | 9.255 | 0.0062 | No | ||
| 3 | NDRG2 | 873 | 7.079 | 0.0009 | No | ||
| 4 | NLGN2 | 1109 | 6.001 | -0.0009 | No | ||
| 5 | RXRB | 1437 | 4.949 | -0.0095 | No | ||
| 6 | PABPC1 | 1592 | 4.530 | -0.0096 | No | ||
| 7 | FANCC | 1629 | 4.432 | -0.0034 | No | ||
| 8 | PSMA1 | 1655 | 4.376 | 0.0033 | No | ||
| 9 | GATA4 | 1834 | 3.993 | 0.0009 | No | ||
| 10 | CD86 | 1882 | 3.880 | 0.0055 | No | ||
| 11 | NUP62 | 2028 | 3.620 | 0.0043 | No | ||
| 12 | STC2 | 2187 | 3.347 | 0.0019 | No | ||
| 13 | STIP1 | 2230 | 3.273 | 0.0056 | No | ||
| 14 | ELOVL6 | 2308 | 3.149 | 0.0072 | No | ||
| 15 | ADAMTS5 | 2498 | 2.912 | 0.0023 | No | ||
| 16 | BFSP1 | 2509 | 2.898 | 0.0070 | No | ||
| 17 | PCDH10 | 2670 | 2.703 | 0.0033 | No | ||
| 18 | CXCL9 | 2926 | 2.453 | -0.0060 | No | ||
| 19 | PLXDC2 | 2997 | 2.390 | -0.0054 | No | ||
| 20 | TCEA2 | 3080 | 2.304 | -0.0056 | No | ||
| 21 | HOXB2 | 3151 | 2.252 | -0.0053 | No | ||
| 22 | SUFU | 3446 | 2.041 | -0.0175 | No | ||
| 23 | FXYD2 | 3528 | 1.990 | -0.0182 | No | ||
| 24 | TNFSF18 | 3547 | 1.981 | -0.0156 | No | ||
| 25 | XPO6 | 3563 | 1.971 | -0.0128 | No | ||
| 26 | RGS3 | 3575 | 1.961 | -0.0098 | No | ||
| 27 | PPP1R13B | 3832 | 1.820 | -0.0203 | No | ||
| 28 | C1QL1 | 3840 | 1.816 | -0.0174 | No | ||
| 29 | PTGS2 | 3930 | 1.769 | -0.0190 | No | ||
| 30 | SIN3A | 4001 | 1.739 | -0.0196 | No | ||
| 31 | MAG | 4067 | 1.706 | -0.0200 | No | ||
| 32 | SLC39A7 | 4111 | 1.685 | -0.0192 | No | ||
| 33 | SYT7 | 4174 | 1.654 | -0.0195 | No | ||
| 34 | TIAL1 | 4197 | 1.643 | -0.0177 | No | ||
| 35 | IL1RN | 4268 | 1.604 | -0.0186 | No | ||
| 36 | DDR1 | 4594 | 1.469 | -0.0335 | No | ||
| 37 | PTHLH | 4608 | 1.464 | -0.0315 | No | ||
| 38 | BDNF | 4697 | 1.430 | -0.0337 | No | ||
| 39 | UBE2I | 4851 | 1.375 | -0.0394 | No | ||
| 40 | RFX5 | 4864 | 1.370 | -0.0376 | No | ||
| 41 | HOXB3 | 4870 | 1.367 | -0.0353 | No | ||
| 42 | FKHL18 | 5071 | 1.296 | -0.0438 | No | ||
| 43 | AMOTL1 | 5119 | 1.280 | -0.0440 | No | ||
| 44 | CYP26A1 | 5228 | 1.247 | -0.0476 | No | ||
| 45 | BCKDK | 5315 | 1.217 | -0.0500 | No | ||
| 46 | CYR61 | 5325 | 1.214 | -0.0483 | No | ||
| 47 | CHD4 | 5615 | 1.122 | -0.0619 | No | ||
| 48 | GPR150 | 5736 | 1.084 | -0.0664 | No | ||
| 49 | CDK6 | 5959 | 1.019 | -0.0766 | No | ||
| 50 | TRIM47 | 6096 | 0.976 | -0.0822 | No | ||
| 51 | HSD3B7 | 6122 | 0.967 | -0.0817 | No | ||
| 52 | PLA1A | 6160 | 0.956 | -0.0820 | No | ||
| 53 | GRIK2 | 6281 | 0.917 | -0.0868 | No | ||
| 54 | NOS1 | 6286 | 0.915 | -0.0854 | No | ||
| 55 | SP6 | 6326 | 0.903 | -0.0858 | No | ||
| 56 | FGF1 | 6361 | 0.893 | -0.0860 | No | ||
| 57 | TMEM27 | 6379 | 0.888 | -0.0853 | No | ||
| 58 | XPO1 | 6402 | 0.881 | -0.0849 | No | ||
| 59 | CLCN1 | 6510 | 0.851 | -0.0891 | No | ||
| 60 | ERN1 | 6533 | 0.844 | -0.0888 | No | ||
| 61 | GRK5 | 6611 | 0.821 | -0.0915 | No | ||
| 62 | WRN | 7015 | 0.720 | -0.1120 | No | ||
| 63 | SLAMF8 | 7494 | 0.590 | -0.1368 | No | ||
| 64 | RGL1 | 7538 | 0.578 | -0.1381 | No | ||
| 65 | IFNB1 | 7668 | 0.547 | -0.1441 | No | ||
| 66 | TCF15 | 7920 | 0.481 | -0.1568 | No | ||
| 67 | CTGF | 7998 | 0.460 | -0.1602 | No | ||
| 68 | PTGES | 8031 | 0.453 | -0.1611 | No | ||
| 69 | SLC6A12 | 8065 | 0.442 | -0.1620 | No | ||
| 70 | VCAM1 | 8195 | 0.411 | -0.1683 | No | ||
| 71 | ARHGEF2 | 8332 | 0.380 | -0.1750 | No | ||
| 72 | GNAO1 | 8399 | 0.362 | -0.1779 | No | ||
| 73 | CHX10 | 8809 | 0.261 | -0.1996 | No | ||
| 74 | TCF1 | 8946 | 0.229 | -0.2065 | No | ||
| 75 | FGF17 | 8973 | 0.220 | -0.2076 | No | ||
| 76 | TSLP | 9008 | 0.214 | -0.2090 | No | ||
| 77 | CGN | 9061 | 0.203 | -0.2114 | No | ||
| 78 | CYBB | 9307 | 0.143 | -0.2245 | No | ||
| 79 | CXCL10 | 9329 | 0.138 | -0.2254 | No | ||
| 80 | CCL5 | 9577 | 0.075 | -0.2386 | No | ||
| 81 | SEC14L2 | 9610 | 0.069 | -0.2402 | No | ||
| 82 | MNT | 9650 | 0.062 | -0.2422 | No | ||
| 83 | IER5 | 9676 | 0.055 | -0.2435 | No | ||
| 84 | PTGIR | 9710 | 0.047 | -0.2452 | No | ||
| 85 | NFKB2 | 9984 | -0.028 | -0.2599 | No | ||
| 86 | KLK9 | 10006 | -0.034 | -0.2610 | No | ||
| 87 | LRP1 | 10267 | -0.095 | -0.2750 | No | ||
| 88 | OGG1 | 10311 | -0.104 | -0.2771 | No | ||
| 89 | TNFSF15 | 10433 | -0.138 | -0.2834 | No | ||
| 90 | RIN2 | 10467 | -0.147 | -0.2849 | No | ||
| 91 | RND1 | 10471 | -0.148 | -0.2848 | No | ||
| 92 | SOX10 | 10727 | -0.217 | -0.2983 | No | ||
| 93 | PURG | 10944 | -0.273 | -0.3095 | No | ||
| 94 | GREM1 | 11027 | -0.296 | -0.3134 | No | ||
| 95 | PDE3B | 11038 | -0.299 | -0.3134 | No | ||
| 96 | EHF | 11135 | -0.327 | -0.3180 | No | ||
| 97 | TOMM20 | 11181 | -0.341 | -0.3198 | No | ||
| 98 | BCL3 | 11717 | -0.485 | -0.3479 | No | ||
| 99 | HOXA11 | 11964 | -0.549 | -0.3602 | No | ||
| 100 | TANK | 12104 | -0.589 | -0.3667 | No | ||
| 101 | CXCL2 | 12340 | -0.662 | -0.3782 | No | ||
| 102 | MSC | 12636 | -0.758 | -0.3928 | No | ||
| 103 | UACA | 12716 | -0.783 | -0.3957 | No | ||
| 104 | SLC16A4 | 12720 | -0.784 | -0.3944 | No | ||
| 105 | PFN1 | 12820 | -0.816 | -0.3983 | No | ||
| 106 | TAL1 | 12905 | -0.841 | -0.4013 | No | ||
| 107 | FOXD2 | 12934 | -0.847 | -0.4013 | No | ||
| 108 | ATF5 | 12975 | -0.863 | -0.4019 | No | ||
| 109 | GABRE | 13287 | -0.977 | -0.4169 | No | ||
| 110 | TNFSF4 | 13397 | -1.016 | -0.4210 | No | ||
| 111 | GJB1 | 13606 | -1.105 | -0.4302 | No | ||
| 112 | CSF2 | 13610 | -1.107 | -0.4284 | No | ||
| 113 | ASCL3 | 13628 | -1.115 | -0.4272 | No | ||
| 114 | HOXB9 | 13694 | -1.131 | -0.4287 | No | ||
| 115 | NFKBIB | 13906 | -1.223 | -0.4379 | No | ||
| 116 | MAP3K8 | 13974 | -1.256 | -0.4392 | No | ||
| 117 | TOP1 | 14010 | -1.275 | -0.4388 | No | ||
| 118 | JAK3 | 14313 | -1.428 | -0.4525 | No | ||
| 119 | CACNA1E | 14331 | -1.437 | -0.4508 | No | ||
| 120 | MLLT6 | 14424 | -1.484 | -0.4531 | No | ||
| 121 | MOBKL2C | 14471 | -1.515 | -0.4528 | No | ||
| 122 | HCFC1 | 14595 | -1.579 | -0.4566 | No | ||
| 123 | PCSK2 | 14596 | -1.580 | -0.4537 | No | ||
| 124 | IL23A | 14650 | -1.608 | -0.4536 | No | ||
| 125 | PAPPA | 14660 | -1.617 | -0.4511 | No | ||
| 126 | ILK | 14906 | -1.749 | -0.4612 | No | ||
| 127 | CYLD | 14912 | -1.752 | -0.4583 | No | ||
| 128 | NLK | 15324 | -2.039 | -0.4768 | No | ||
| 129 | BNC2 | 15533 | -2.244 | -0.4840 | No | ||
| 130 | ATP1B3 | 15573 | -2.278 | -0.4819 | No | ||
| 131 | ZADH2 | 15669 | -2.373 | -0.4827 | No | ||
| 132 | SEMA3B | 15715 | -2.419 | -0.4807 | No | ||
| 133 | SLC11A2 | 15773 | -2.475 | -0.4793 | No | ||
| 134 | NR2F2 | 15791 | -2.488 | -0.4756 | No | ||
| 135 | NFAT5 | 16018 | -2.763 | -0.4828 | No | ||
| 136 | BCOR | 16289 | -3.166 | -0.4917 | Yes | ||
| 137 | ICAM1 | 16314 | -3.203 | -0.4871 | Yes | ||
| 138 | PDE4C | 16337 | -3.245 | -0.4823 | Yes | ||
| 139 | AP1S2 | 16346 | -3.260 | -0.4768 | Yes | ||
| 140 | REL | 16362 | -3.269 | -0.4716 | Yes | ||
| 141 | PRKCD | 16381 | -3.304 | -0.4665 | Yes | ||
| 142 | CPLX2 | 16390 | -3.317 | -0.4609 | Yes | ||
| 143 | RYBP | 16395 | -3.335 | -0.4550 | Yes | ||
| 144 | RAI1 | 16496 | -3.548 | -0.4539 | Yes | ||
| 145 | ASH1L | 16621 | -3.796 | -0.4536 | Yes | ||
| 146 | TNIP1 | 16627 | -3.816 | -0.4469 | Yes | ||
| 147 | HOXA7 | 16690 | -3.928 | -0.4431 | Yes | ||
| 148 | PPARBP | 16743 | -4.058 | -0.4384 | Yes | ||
| 149 | PLAU | 16814 | -4.227 | -0.4345 | Yes | ||
| 150 | WNT10A | 16853 | -4.299 | -0.4287 | Yes | ||
| 151 | ZHX2 | 16890 | -4.381 | -0.4226 | Yes | ||
| 152 | GBA2 | 16906 | -4.422 | -0.4153 | Yes | ||
| 153 | CDC14A | 16908 | -4.437 | -0.4072 | Yes | ||
| 154 | DNAJB5 | 17042 | -4.686 | -0.4058 | Yes | ||
| 155 | RAP2C | 17082 | -4.821 | -0.3991 | Yes | ||
| 156 | ACTR1A | 17162 | -5.066 | -0.3941 | Yes | ||
| 157 | TRIB2 | 17257 | -5.354 | -0.3893 | Yes | ||
| 158 | CD83 | 17275 | -5.409 | -0.3803 | Yes | ||
| 159 | IL2RA | 17295 | -5.472 | -0.3713 | Yes | ||
| 160 | STAT6 | 17308 | -5.487 | -0.3619 | Yes | ||
| 161 | PAFAH1B3 | 17512 | -6.072 | -0.3618 | Yes | ||
| 162 | NSD1 | 17558 | -6.273 | -0.3527 | Yes | ||
| 163 | ARPC5 | 17596 | -6.399 | -0.3430 | Yes | ||
| 164 | P2RY10 | 17601 | -6.419 | -0.3314 | Yes | ||
| 165 | PTPRJ | 17664 | -6.637 | -0.3226 | Yes | ||
| 166 | TGFB1 | 17717 | -6.795 | -0.3130 | Yes | ||
| 167 | RUNX1 | 17822 | -7.209 | -0.3054 | Yes | ||
| 168 | NFKBIA | 17884 | -7.579 | -0.2948 | Yes | ||
| 169 | LTB | 17951 | -7.948 | -0.2838 | Yes | ||
| 170 | ATP1B1 | 17985 | -8.207 | -0.2705 | Yes | ||
| 171 | LASP1 | 18052 | -8.575 | -0.2584 | Yes | ||
| 172 | BMF | 18150 | -9.381 | -0.2464 | Yes | ||
| 173 | BLR1 | 18153 | -9.416 | -0.2292 | Yes | ||
| 174 | LMO4 | 18158 | -9.497 | -0.2120 | Yes | ||
| 175 | EPHA2 | 18212 | -10.078 | -0.1964 | Yes | ||
| 176 | SATB1 | 18248 | -10.370 | -0.1793 | Yes | ||
| 177 | SAMSN1 | 18381 | -12.404 | -0.1637 | Yes | ||
| 178 | SOX5 | 18526 | -18.118 | -0.1383 | Yes | ||
| 179 | IL4I1 | 18563 | -26.226 | -0.0921 | Yes | ||
| 180 | BHLHB2 | 18597 | -51.746 | 0.0010 | Yes |