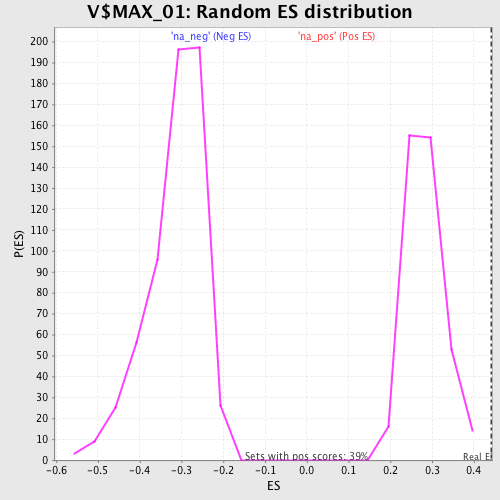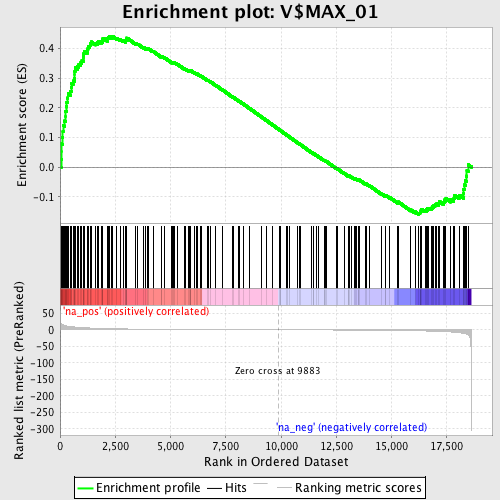
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MAX_01 |
| Enrichment Score (ES) | 0.441193 |
| Normalized Enrichment Score (NES) | 1.5618737 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.048827153 |
| FWER p-Value | 0.831 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RCL1 | 46 | 19.200 | 0.0258 | Yes | ||
| 2 | ACY1 | 57 | 18.355 | 0.0523 | Yes | ||
| 3 | TIMM8A | 60 | 18.225 | 0.0791 | Yes | ||
| 4 | FKBP11 | 97 | 15.642 | 0.1002 | Yes | ||
| 5 | AATF | 128 | 14.571 | 0.1200 | Yes | ||
| 6 | CAD | 139 | 14.296 | 0.1405 | Yes | ||
| 7 | NUDC | 206 | 12.580 | 0.1555 | Yes | ||
| 8 | ATF4 | 248 | 11.928 | 0.1709 | Yes | ||
| 9 | SUCLG2 | 253 | 11.870 | 0.1881 | Yes | ||
| 10 | OPRS1 | 289 | 11.463 | 0.2031 | Yes | ||
| 11 | SUPT16H | 302 | 11.270 | 0.2191 | Yes | ||
| 12 | QTRT1 | 342 | 10.813 | 0.2329 | Yes | ||
| 13 | BAX | 357 | 10.616 | 0.2478 | Yes | ||
| 14 | TIMM10 | 474 | 9.631 | 0.2557 | Yes | ||
| 15 | PPRC1 | 502 | 9.457 | 0.2682 | Yes | ||
| 16 | RPS2 | 526 | 9.310 | 0.2807 | Yes | ||
| 17 | MTHFD1 | 589 | 8.792 | 0.2902 | Yes | ||
| 18 | AKAP1 | 648 | 8.392 | 0.2995 | Yes | ||
| 19 | EIF4E | 663 | 8.322 | 0.3110 | Yes | ||
| 20 | PA2G4 | 666 | 8.313 | 0.3231 | Yes | ||
| 21 | SYNCRIP | 679 | 8.203 | 0.3346 | Yes | ||
| 22 | AMPD2 | 789 | 7.536 | 0.3398 | Yes | ||
| 23 | MRPL40 | 848 | 7.236 | 0.3473 | Yes | ||
| 24 | QTRTD1 | 939 | 6.783 | 0.3524 | Yes | ||
| 25 | ANAPC13 | 988 | 6.524 | 0.3594 | Yes | ||
| 26 | MTCH2 | 1048 | 6.294 | 0.3655 | Yes | ||
| 27 | CLN3 | 1052 | 6.281 | 0.3746 | Yes | ||
| 28 | EIF3S10 | 1075 | 6.154 | 0.3825 | Yes | ||
| 29 | PICALM | 1096 | 6.067 | 0.3903 | Yes | ||
| 30 | GTF2H1 | 1223 | 5.646 | 0.3918 | Yes | ||
| 31 | EEF1B2 | 1259 | 5.530 | 0.3981 | Yes | ||
| 32 | MRPL27 | 1265 | 5.512 | 0.4059 | Yes | ||
| 33 | FEN1 | 1352 | 5.246 | 0.4090 | Yes | ||
| 34 | PSME3 | 1362 | 5.209 | 0.4162 | Yes | ||
| 35 | PRPS1 | 1398 | 5.067 | 0.4217 | Yes | ||
| 36 | PABPC1 | 1592 | 4.530 | 0.4180 | Yes | ||
| 37 | ATP6V1C1 | 1688 | 4.322 | 0.4192 | Yes | ||
| 38 | PPP1R3C | 1747 | 4.195 | 0.4222 | Yes | ||
| 39 | NFX1 | 1859 | 3.927 | 0.4220 | Yes | ||
| 40 | HNRPA1 | 1915 | 3.826 | 0.4246 | Yes | ||
| 41 | MAT2A | 1931 | 3.792 | 0.4294 | Yes | ||
| 42 | HIRA | 1938 | 3.779 | 0.4347 | Yes | ||
| 43 | PFDN2 | 2134 | 3.440 | 0.4292 | Yes | ||
| 44 | PTGES2 | 2149 | 3.406 | 0.4334 | Yes | ||
| 45 | SYT3 | 2181 | 3.355 | 0.4367 | Yes | ||
| 46 | HMGA1 | 2217 | 3.288 | 0.4396 | Yes | ||
| 47 | DHX35 | 2330 | 3.111 | 0.4381 | Yes | ||
| 48 | CPT1A | 2358 | 3.060 | 0.4412 | Yes | ||
| 49 | TGFB2 | 2556 | 2.830 | 0.4347 | No | ||
| 50 | ASNA1 | 2736 | 2.623 | 0.4288 | No | ||
| 51 | ATOH8 | 2862 | 2.510 | 0.4258 | No | ||
| 52 | TFRC | 2957 | 2.430 | 0.4243 | No | ||
| 53 | ILF3 | 2971 | 2.413 | 0.4271 | No | ||
| 54 | DDX3X | 2995 | 2.391 | 0.4294 | No | ||
| 55 | SRP72 | 3020 | 2.369 | 0.4316 | No | ||
| 56 | EIF4B | 3024 | 2.367 | 0.4349 | No | ||
| 57 | NPM1 | 3426 | 2.056 | 0.4162 | No | ||
| 58 | CFL1 | 3515 | 1.999 | 0.4144 | No | ||
| 59 | EN1 | 3767 | 1.848 | 0.4035 | No | ||
| 60 | DSCAM | 3885 | 1.792 | 0.3998 | No | ||
| 61 | STMN1 | 3958 | 1.757 | 0.3985 | No | ||
| 62 | RAB3IL1 | 4004 | 1.738 | 0.3986 | No | ||
| 63 | LZTS2 | 4205 | 1.638 | 0.3902 | No | ||
| 64 | POGK | 4576 | 1.479 | 0.3723 | No | ||
| 65 | CENTB5 | 4590 | 1.472 | 0.3738 | No | ||
| 66 | BOK | 4741 | 1.414 | 0.3677 | No | ||
| 67 | HOXD10 | 5058 | 1.300 | 0.3525 | No | ||
| 68 | TCOF1 | 5108 | 1.286 | 0.3517 | No | ||
| 69 | KCNE4 | 5126 | 1.279 | 0.3527 | No | ||
| 70 | UBE4B | 5178 | 1.262 | 0.3518 | No | ||
| 71 | MEOX2 | 5306 | 1.219 | 0.3467 | No | ||
| 72 | CHD4 | 5615 | 1.122 | 0.3317 | No | ||
| 73 | DUSP7 | 5668 | 1.102 | 0.3305 | No | ||
| 74 | NDUFS1 | 5802 | 1.062 | 0.3248 | No | ||
| 75 | OPRD1 | 5854 | 1.047 | 0.3236 | No | ||
| 76 | IPO13 | 5879 | 1.039 | 0.3238 | No | ||
| 77 | ELAVL3 | 5893 | 1.035 | 0.3247 | No | ||
| 78 | ADAMTS17 | 5901 | 1.033 | 0.3258 | No | ||
| 79 | CCAR1 | 6077 | 0.981 | 0.3178 | No | ||
| 80 | SSR1 | 6182 | 0.949 | 0.3135 | No | ||
| 81 | RUNX2 | 6201 | 0.942 | 0.3139 | No | ||
| 82 | DVL2 | 6338 | 0.900 | 0.3079 | No | ||
| 83 | XPO1 | 6402 | 0.881 | 0.3058 | No | ||
| 84 | SNCAIP | 6672 | 0.806 | 0.2924 | No | ||
| 85 | RAI14 | 6699 | 0.798 | 0.2921 | No | ||
| 86 | TCERG1 | 6793 | 0.776 | 0.2882 | No | ||
| 87 | ODC1 | 7047 | 0.711 | 0.2756 | No | ||
| 88 | HOXB5 | 7359 | 0.628 | 0.2596 | No | ||
| 89 | GUCY1A2 | 7796 | 0.514 | 0.2368 | No | ||
| 90 | PCDHA10 | 7868 | 0.492 | 0.2336 | No | ||
| 91 | ZCCHC7 | 8077 | 0.438 | 0.2230 | No | ||
| 92 | SEPT3 | 8127 | 0.426 | 0.2210 | No | ||
| 93 | CDK5R1 | 8277 | 0.392 | 0.2135 | No | ||
| 94 | LAMP1 | 8581 | 0.317 | 0.1975 | No | ||
| 95 | TESK2 | 9115 | 0.191 | 0.1689 | No | ||
| 96 | SMAD2 | 9357 | 0.129 | 0.1560 | No | ||
| 97 | GATA5 | 9592 | 0.072 | 0.1435 | No | ||
| 98 | SUMF1 | 9949 | -0.018 | 0.1242 | No | ||
| 99 | NPTX1 | 9970 | -0.024 | 0.1231 | No | ||
| 100 | NEUROD1 | 9994 | -0.031 | 0.1219 | No | ||
| 101 | LZTFL1 | 10240 | -0.087 | 0.1088 | No | ||
| 102 | COPZ1 | 10302 | -0.102 | 0.1056 | No | ||
| 103 | SLC26A2 | 10363 | -0.120 | 0.1026 | No | ||
| 104 | PTMA | 10754 | -0.223 | 0.0818 | No | ||
| 105 | PRKCH | 10831 | -0.243 | 0.0780 | No | ||
| 106 | BRDT | 10875 | -0.257 | 0.0760 | No | ||
| 107 | ZBTB10 | 11380 | -0.391 | 0.0493 | No | ||
| 108 | BMP6 | 11477 | -0.419 | 0.0447 | No | ||
| 109 | LIN28 | 11478 | -0.419 | 0.0453 | No | ||
| 110 | HOXB7 | 11617 | -0.461 | 0.0385 | No | ||
| 111 | PER1 | 11715 | -0.484 | 0.0340 | No | ||
| 112 | TOPORS | 11953 | -0.547 | 0.0219 | No | ||
| 113 | HOXA11 | 11964 | -0.549 | 0.0222 | No | ||
| 114 | FOXD3 | 12009 | -0.560 | 0.0206 | No | ||
| 115 | UBE2B | 12066 | -0.577 | 0.0185 | No | ||
| 116 | FGF14 | 12487 | -0.707 | -0.0033 | No | ||
| 117 | HOXC5 | 12562 | -0.733 | -0.0062 | No | ||
| 118 | SNX5 | 12856 | -0.826 | -0.0209 | No | ||
| 119 | UBXD3 | 13053 | -0.896 | -0.0302 | No | ||
| 120 | ARMET | 13057 | -0.897 | -0.0290 | No | ||
| 121 | NRIP3 | 13089 | -0.907 | -0.0293 | No | ||
| 122 | SLC38A5 | 13186 | -0.943 | -0.0332 | No | ||
| 123 | GGN | 13330 | -0.993 | -0.0394 | No | ||
| 124 | AKAP12 | 13353 | -1.001 | -0.0392 | No | ||
| 125 | CBX5 | 13421 | -1.026 | -0.0413 | No | ||
| 126 | DNMT3A | 13484 | -1.060 | -0.0431 | No | ||
| 127 | SLC1A7 | 13492 | -1.064 | -0.0419 | No | ||
| 128 | VPS16 | 13542 | -1.083 | -0.0430 | No | ||
| 129 | TAF6L | 13802 | -1.183 | -0.0553 | No | ||
| 130 | SNTB2 | 13855 | -1.203 | -0.0563 | No | ||
| 131 | TOP1 | 14010 | -1.275 | -0.0628 | No | ||
| 132 | ATP6V1A | 14557 | -1.557 | -0.0901 | No | ||
| 133 | PDK2 | 14720 | -1.649 | -0.0964 | No | ||
| 134 | IRS4 | 14736 | -1.655 | -0.0948 | No | ||
| 135 | GNAS | 14925 | -1.757 | -0.1024 | No | ||
| 136 | COMMD3 | 15284 | -2.010 | -0.1188 | No | ||
| 137 | MAX | 15307 | -2.029 | -0.1170 | No | ||
| 138 | SLC31A2 | 15874 | -2.583 | -0.1439 | No | ||
| 139 | NIT1 | 16071 | -2.831 | -0.1504 | No | ||
| 140 | ZFYVE26 | 16239 | -3.077 | -0.1549 | No | ||
| 141 | BCOR | 16289 | -3.166 | -0.1529 | No | ||
| 142 | GPX1 | 16313 | -3.203 | -0.1494 | No | ||
| 143 | SIRT1 | 16332 | -3.238 | -0.1456 | No | ||
| 144 | AP1S2 | 16346 | -3.260 | -0.1415 | No | ||
| 145 | GNB2 | 16519 | -3.605 | -0.1455 | No | ||
| 146 | USP15 | 16587 | -3.735 | -0.1437 | No | ||
| 147 | PPM1A | 16623 | -3.801 | -0.1399 | No | ||
| 148 | HOXA7 | 16690 | -3.928 | -0.1377 | No | ||
| 149 | ZNF318 | 16802 | -4.206 | -0.1376 | No | ||
| 150 | SLC38A2 | 16844 | -4.285 | -0.1335 | No | ||
| 151 | EIF2C2 | 16911 | -4.445 | -0.1305 | No | ||
| 152 | MXD4 | 16968 | -4.544 | -0.1268 | No | ||
| 153 | DNAJB5 | 17042 | -4.686 | -0.1239 | No | ||
| 154 | ALDH6A1 | 17130 | -4.971 | -0.1213 | No | ||
| 155 | TCF4 | 17181 | -5.117 | -0.1164 | No | ||
| 156 | BCAS3 | 17364 | -5.682 | -0.1179 | No | ||
| 157 | DUSP1 | 17384 | -5.737 | -0.1105 | No | ||
| 158 | NOTCH1 | 17461 | -5.950 | -0.1059 | No | ||
| 159 | PLCG2 | 17677 | -6.671 | -0.1077 | No | ||
| 160 | CD164 | 17823 | -7.211 | -0.1049 | No | ||
| 161 | KBTBD2 | 17854 | -7.427 | -0.0956 | No | ||
| 162 | ETV1 | 18059 | -8.622 | -0.0939 | No | ||
| 163 | ATF7IP | 18257 | -10.399 | -0.0893 | No | ||
| 164 | EPC1 | 18267 | -10.496 | -0.0743 | No | ||
| 165 | HHEX | 18291 | -10.672 | -0.0598 | No | ||
| 166 | CSK | 18344 | -11.494 | -0.0457 | No | ||
| 167 | GJA1 | 18398 | -12.744 | -0.0298 | No | ||
| 168 | ISGF3G | 18414 | -13.083 | -0.0113 | No | ||
| 169 | HBP1 | 18472 | -15.082 | 0.0078 | No |