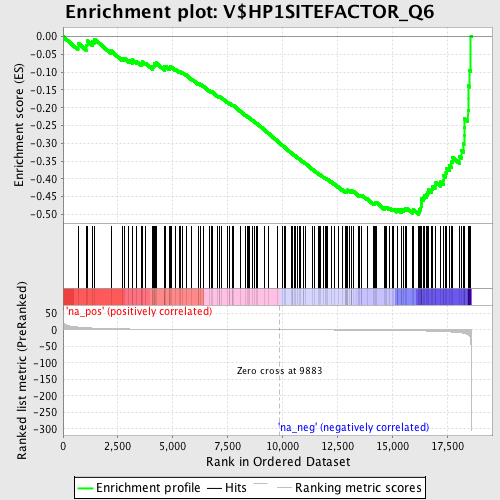
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$HP1SITEFACTOR_Q6 |
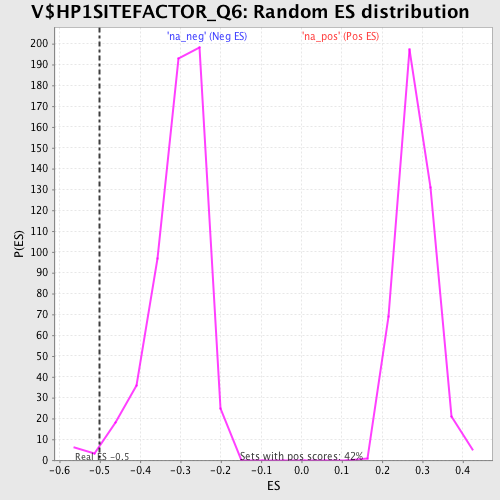
| Enrichment Score (ES) | -0.5002125 |
| Normalized Enrichment Score (NES) | -1.6349269 |
| Nominal p-value | 0.013888889 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SYNCRIP | 679 | 8.203 | -0.0181 | No | ||
| 2 | MTCH2 | 1048 | 6.294 | -0.0236 | No | ||
| 3 | MRPL28 | 1089 | 6.109 | -0.0119 | No | ||
| 4 | LUC7L | 1349 | 5.250 | -0.0139 | No | ||
| 5 | SFRS2 | 1426 | 4.971 | -0.0067 | No | ||
| 6 | NR5A2 | 2184 | 3.351 | -0.0400 | No | ||
| 7 | ADAM28 | 2692 | 2.675 | -0.0614 | No | ||
| 8 | GCNT2 | 2780 | 2.577 | -0.0602 | No | ||
| 9 | CACNA2D3 | 2994 | 2.394 | -0.0663 | No | ||
| 10 | HOXA9 | 3152 | 2.250 | -0.0697 | No | ||
| 11 | PRICKLE1 | 3160 | 2.243 | -0.0649 | No | ||
| 12 | KIF1B | 3355 | 2.105 | -0.0707 | No | ||
| 13 | RGS3 | 3575 | 1.961 | -0.0780 | No | ||
| 14 | PAPD1 | 3593 | 1.949 | -0.0745 | No | ||
| 15 | ADAM11 | 3607 | 1.942 | -0.0708 | No | ||
| 16 | ACCN4 | 3771 | 1.847 | -0.0754 | No | ||
| 17 | CDC2L2 | 4078 | 1.699 | -0.0881 | No | ||
| 18 | RDH10 | 4081 | 1.698 | -0.0843 | No | ||
| 19 | RBP4 | 4136 | 1.671 | -0.0834 | No | ||
| 20 | VLDLR | 4148 | 1.667 | -0.0802 | No | ||
| 21 | CHRDL1 | 4153 | 1.666 | -0.0766 | No | ||
| 22 | BHLHB3 | 4219 | 1.627 | -0.0765 | No | ||
| 23 | NCALD | 4237 | 1.618 | -0.0737 | No | ||
| 24 | BARHL1 | 4618 | 1.462 | -0.0909 | No | ||
| 25 | SOX2 | 4628 | 1.459 | -0.0881 | No | ||
| 26 | PHF16 | 4634 | 1.457 | -0.0850 | No | ||
| 27 | EML4 | 4682 | 1.437 | -0.0843 | No | ||
| 28 | FBN2 | 4852 | 1.375 | -0.0903 | No | ||
| 29 | HOXB3 | 4870 | 1.367 | -0.0881 | No | ||
| 30 | GDF8 | 4871 | 1.367 | -0.0850 | No | ||
| 31 | LMNA | 4935 | 1.343 | -0.0853 | No | ||
| 32 | PPP1CB | 5113 | 1.284 | -0.0920 | No | ||
| 33 | ZIC1 | 5285 | 1.225 | -0.0985 | No | ||
| 34 | SEZ6 | 5327 | 1.214 | -0.0979 | No | ||
| 35 | GPR27 | 5454 | 1.172 | -0.1021 | No | ||
| 36 | MYH2 | 5604 | 1.125 | -0.1076 | No | ||
| 37 | EVI1 | 5872 | 1.043 | -0.1197 | No | ||
| 38 | ATP6V1E2 | 6148 | 0.960 | -0.1324 | No | ||
| 39 | PROK2 | 6161 | 0.956 | -0.1308 | No | ||
| 40 | AFP | 6267 | 0.920 | -0.1344 | No | ||
| 41 | XPO1 | 6402 | 0.881 | -0.1397 | No | ||
| 42 | SNCAIP | 6672 | 0.806 | -0.1524 | No | ||
| 43 | MYCBP | 6774 | 0.781 | -0.1561 | No | ||
| 44 | LHX6 | 6795 | 0.775 | -0.1554 | No | ||
| 45 | STC1 | 7040 | 0.714 | -0.1670 | No | ||
| 46 | TYRO3 | 7108 | 0.696 | -0.1691 | No | ||
| 47 | CNTFR | 7130 | 0.689 | -0.1686 | No | ||
| 48 | ANPEP | 7207 | 0.671 | -0.1712 | No | ||
| 49 | FMO3 | 7488 | 0.592 | -0.1850 | No | ||
| 50 | CA4 | 7588 | 0.567 | -0.1891 | No | ||
| 51 | HSD17B2 | 7590 | 0.567 | -0.1879 | No | ||
| 52 | IBRDC2 | 7714 | 0.537 | -0.1933 | No | ||
| 53 | IGF1 | 7738 | 0.530 | -0.1933 | No | ||
| 54 | NPAS3 | 7757 | 0.524 | -0.1931 | No | ||
| 55 | ATP2B3 | 8072 | 0.438 | -0.2091 | No | ||
| 56 | USP13 | 8326 | 0.381 | -0.2220 | No | ||
| 57 | PJA1 | 8407 | 0.361 | -0.2255 | No | ||
| 58 | GPC3 | 8430 | 0.353 | -0.2259 | No | ||
| 59 | HOXD3 | 8474 | 0.343 | -0.2274 | No | ||
| 60 | NFIX | 8641 | 0.303 | -0.2357 | No | ||
| 61 | POU4F1 | 8733 | 0.283 | -0.2400 | No | ||
| 62 | LHX1 | 8814 | 0.260 | -0.2437 | No | ||
| 63 | BMP5 | 8852 | 0.252 | -0.2452 | No | ||
| 64 | CS | 9180 | 0.175 | -0.2625 | No | ||
| 65 | DLX1 | 9366 | 0.128 | -0.2722 | No | ||
| 66 | GPR110 | 9767 | 0.031 | -0.2938 | No | ||
| 67 | NOG | 9992 | -0.031 | -0.3059 | No | ||
| 68 | CRYGD | 10088 | -0.053 | -0.3109 | No | ||
| 69 | DRD3 | 10126 | -0.062 | -0.3128 | No | ||
| 70 | HOXA4 | 10386 | -0.126 | -0.3265 | No | ||
| 71 | FAP | 10439 | -0.140 | -0.3290 | No | ||
| 72 | RIN2 | 10467 | -0.147 | -0.3302 | No | ||
| 73 | RCOR1 | 10551 | -0.166 | -0.3343 | No | ||
| 74 | PCDHA7 | 10595 | -0.178 | -0.3362 | No | ||
| 75 | SCHIP1 | 10667 | -0.200 | -0.3396 | No | ||
| 76 | DIO2 | 10753 | -0.222 | -0.3437 | No | ||
| 77 | SPARC | 10808 | -0.237 | -0.3461 | No | ||
| 78 | EIF4G2 | 10936 | -0.272 | -0.3523 | No | ||
| 79 | LEMD1 | 10976 | -0.280 | -0.3538 | No | ||
| 80 | JAG1 | 11026 | -0.296 | -0.3558 | No | ||
| 81 | ZBTB10 | 11380 | -0.391 | -0.3740 | No | ||
| 82 | AXL | 11455 | -0.414 | -0.3771 | No | ||
| 83 | HOXB7 | 11617 | -0.461 | -0.3848 | No | ||
| 84 | FLRT1 | 11701 | -0.482 | -0.3882 | No | ||
| 85 | CSF3 | 11725 | -0.488 | -0.3883 | No | ||
| 86 | PITX2 | 11869 | -0.524 | -0.3949 | No | ||
| 87 | HOXA11 | 11964 | -0.549 | -0.3987 | No | ||
| 88 | FOXD3 | 12009 | -0.560 | -0.3998 | No | ||
| 89 | POLK | 12070 | -0.578 | -0.4017 | No | ||
| 90 | OFCC1 | 12214 | -0.626 | -0.4080 | No | ||
| 91 | FBXL19 | 12359 | -0.668 | -0.4143 | No | ||
| 92 | CALCRL | 12538 | -0.724 | -0.4223 | No | ||
| 93 | ESRRG | 12732 | -0.787 | -0.4310 | No | ||
| 94 | PIK3R1 | 12882 | -0.833 | -0.4371 | No | ||
| 95 | TSHB | 12887 | -0.835 | -0.4355 | No | ||
| 96 | ACRV1 | 12935 | -0.848 | -0.4361 | No | ||
| 97 | PARD6A | 12936 | -0.849 | -0.4341 | No | ||
| 98 | DMD | 12944 | -0.850 | -0.4326 | No | ||
| 99 | DEPDC2 | 12956 | -0.854 | -0.4312 | No | ||
| 100 | UBXD3 | 13053 | -0.896 | -0.4344 | No | ||
| 101 | SFXN5 | 13124 | -0.922 | -0.4361 | No | ||
| 102 | NCOR1 | 13128 | -0.926 | -0.4341 | No | ||
| 103 | PLS3 | 13137 | -0.928 | -0.4324 | No | ||
| 104 | MAPK10 | 13249 | -0.964 | -0.4362 | No | ||
| 105 | RHOQ | 13483 | -1.060 | -0.4464 | No | ||
| 106 | TBX2 | 13511 | -1.072 | -0.4455 | No | ||
| 107 | HAPLN2 | 13588 | -1.101 | -0.4471 | No | ||
| 108 | A2BP1 | 13618 | -1.110 | -0.4461 | No | ||
| 109 | GPR156 | 13859 | -1.204 | -0.4564 | No | ||
| 110 | STAG2 | 14159 | -1.349 | -0.4695 | No | ||
| 111 | MAP4K4 | 14202 | -1.367 | -0.4686 | No | ||
| 112 | H2AFY | 14218 | -1.376 | -0.4663 | No | ||
| 113 | POU4F2 | 14290 | -1.414 | -0.4669 | No | ||
| 114 | NRXN3 | 14648 | -1.606 | -0.4826 | No | ||
| 115 | POU3F3 | 14687 | -1.630 | -0.4809 | No | ||
| 116 | LRFN5 | 14750 | -1.664 | -0.4805 | No | ||
| 117 | VAMP3 | 14867 | -1.728 | -0.4828 | No | ||
| 118 | FOXA2 | 14993 | -1.807 | -0.4855 | No | ||
| 119 | DPYSL2 | 15060 | -1.859 | -0.4848 | No | ||
| 120 | TNNC1 | 15217 | -1.957 | -0.4888 | No | ||
| 121 | PUM2 | 15247 | -1.974 | -0.4859 | No | ||
| 122 | HIVEP3 | 15426 | -2.141 | -0.4906 | No | ||
| 123 | AP1G1 | 15433 | -2.145 | -0.4861 | No | ||
| 124 | BNC2 | 15533 | -2.244 | -0.4863 | No | ||
| 125 | PRKAG1 | 15583 | -2.286 | -0.4837 | No | ||
| 126 | ZADH2 | 15669 | -2.373 | -0.4829 | No | ||
| 127 | FOXP2 | 15917 | -2.636 | -0.4903 | No | ||
| 128 | TGFB3 | 15969 | -2.702 | -0.4869 | No | ||
| 129 | TNKS1BP1 | 16216 | -3.033 | -0.4933 | Yes | ||
| 130 | NOSIP | 16230 | -3.063 | -0.4870 | Yes | ||
| 131 | CYFIP2 | 16272 | -3.137 | -0.4821 | Yes | ||
| 132 | H3F3B | 16311 | -3.201 | -0.4768 | Yes | ||
| 133 | RAB3IP | 16318 | -3.207 | -0.4698 | Yes | ||
| 134 | COL4A3BP | 16334 | -3.240 | -0.4632 | Yes | ||
| 135 | AP1S2 | 16346 | -3.260 | -0.4564 | Yes | ||
| 136 | TAPBP | 16417 | -3.381 | -0.4525 | Yes | ||
| 137 | DMTF1 | 16467 | -3.487 | -0.4471 | Yes | ||
| 138 | CASP8 | 16569 | -3.698 | -0.4442 | Yes | ||
| 139 | CDKL5 | 16605 | -3.769 | -0.4375 | Yes | ||
| 140 | ELF4 | 16652 | -3.860 | -0.4311 | Yes | ||
| 141 | POU2F1 | 16806 | -4.210 | -0.4298 | Yes | ||
| 142 | CHD2 | 16826 | -4.251 | -0.4212 | Yes | ||
| 143 | MEIS1 | 16952 | -4.525 | -0.4176 | Yes | ||
| 144 | MAP2K6 | 16991 | -4.589 | -0.4092 | Yes | ||
| 145 | HDAC9 | 17200 | -5.177 | -0.4086 | Yes | ||
| 146 | PRKACB | 17345 | -5.621 | -0.4036 | Yes | ||
| 147 | CITED2 | 17354 | -5.650 | -0.3911 | Yes | ||
| 148 | CTCF | 17449 | -5.920 | -0.3827 | Yes | ||
| 149 | SMAD1 | 17482 | -5.985 | -0.3708 | Yes | ||
| 150 | P2RY10 | 17601 | -6.419 | -0.3625 | Yes | ||
| 151 | BCL11A | 17700 | -6.753 | -0.3524 | Yes | ||
| 152 | MBNL1 | 17740 | -6.866 | -0.3388 | Yes | ||
| 153 | MBNL2 | 18065 | -8.653 | -0.3366 | Yes | ||
| 154 | LMO4 | 18158 | -9.497 | -0.3199 | Yes | ||
| 155 | SCML4 | 18252 | -10.384 | -0.3013 | Yes | ||
| 156 | SOX4 | 18273 | -10.557 | -0.2783 | Yes | ||
| 157 | ZFP36L1 | 18278 | -10.574 | -0.2543 | Yes | ||
| 158 | HHEX | 18291 | -10.672 | -0.2306 | Yes | ||
| 159 | BCL6 | 18453 | -14.443 | -0.2064 | Yes | ||
| 160 | TLE4 | 18471 | -15.081 | -0.1729 | Yes | ||
| 161 | HBP1 | 18472 | -15.082 | -0.1384 | Yes | ||
| 162 | DUSP10 | 18536 | -20.147 | -0.0958 | Yes | ||
| 163 | LRRN3 | 18589 | -43.847 | 0.0015 | Yes |