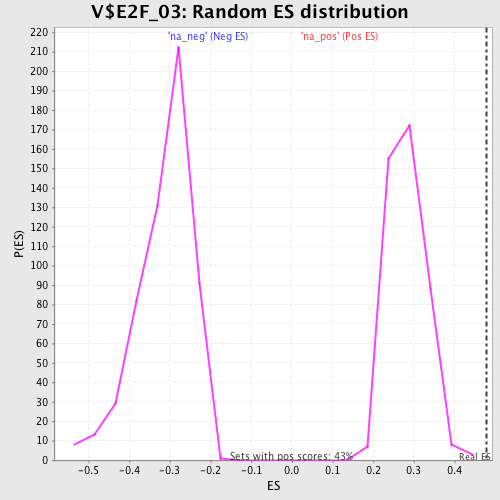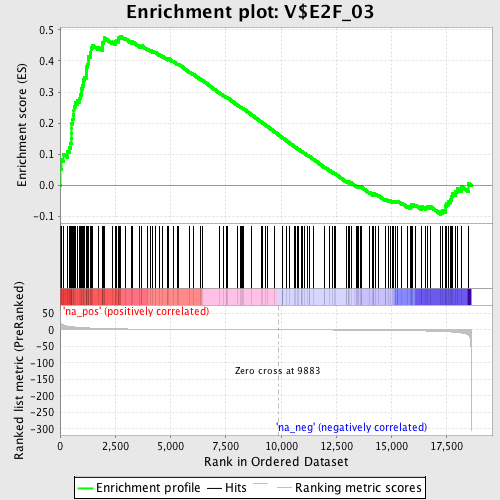
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_03 |
| Enrichment Score (ES) | 0.47952685 |
| Normalized Enrichment Score (NES) | 1.6962667 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.052988928 |
| FWER p-Value | 0.309 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MCM6 | 9 | 30.323 | 0.0521 | Yes | ||
| 2 | RANBP1 | 55 | 18.855 | 0.0824 | Yes | ||
| 3 | AK2 | 153 | 13.651 | 0.1009 | Yes | ||
| 4 | UNG | 340 | 10.863 | 0.1096 | Yes | ||
| 5 | EIF5A | 422 | 10.060 | 0.1227 | Yes | ||
| 6 | RAD51 | 478 | 9.610 | 0.1364 | Yes | ||
| 7 | PPRC1 | 502 | 9.457 | 0.1516 | Yes | ||
| 8 | CDC6 | 518 | 9.366 | 0.1670 | Yes | ||
| 9 | ORC1L | 524 | 9.323 | 0.1829 | Yes | ||
| 10 | RPA2 | 529 | 9.268 | 0.1988 | Yes | ||
| 11 | TCP1 | 561 | 9.012 | 0.2128 | Yes | ||
| 12 | CDCA7 | 587 | 8.803 | 0.2267 | Yes | ||
| 13 | MCM7 | 624 | 8.576 | 0.2396 | Yes | ||
| 14 | MRPL18 | 629 | 8.550 | 0.2543 | Yes | ||
| 15 | NR4A2 | 677 | 8.211 | 0.2660 | Yes | ||
| 16 | MCM3 | 785 | 7.561 | 0.2733 | Yes | ||
| 17 | FKBP5 | 889 | 6.987 | 0.2798 | Yes | ||
| 18 | NUTF2 | 909 | 6.922 | 0.2908 | Yes | ||
| 19 | CDC25A | 963 | 6.668 | 0.2995 | Yes | ||
| 20 | NASP | 969 | 6.607 | 0.3107 | Yes | ||
| 21 | POLA2 | 997 | 6.482 | 0.3205 | Yes | ||
| 22 | LIG1 | 1042 | 6.323 | 0.3291 | Yes | ||
| 23 | MCM4 | 1059 | 6.251 | 0.3391 | Yes | ||
| 24 | PRKCSH | 1099 | 6.051 | 0.3475 | Yes | ||
| 25 | CDC45L | 1175 | 5.835 | 0.3535 | Yes | ||
| 26 | KCNA6 | 1188 | 5.799 | 0.3630 | Yes | ||
| 27 | KCND2 | 1204 | 5.719 | 0.3721 | Yes | ||
| 28 | SUV39H1 | 1211 | 5.672 | 0.3816 | Yes | ||
| 29 | ANP32A | 1241 | 5.582 | 0.3897 | Yes | ||
| 30 | MCM2 | 1266 | 5.511 | 0.3980 | Yes | ||
| 31 | GMNN | 1272 | 5.493 | 0.4072 | Yes | ||
| 32 | SMAD6 | 1293 | 5.438 | 0.4156 | Yes | ||
| 33 | RFC1 | 1375 | 5.143 | 0.4201 | Yes | ||
| 34 | KNTC1 | 1386 | 5.099 | 0.4284 | Yes | ||
| 35 | PRPS1 | 1398 | 5.067 | 0.4366 | Yes | ||
| 36 | SFRS2 | 1426 | 4.971 | 0.4438 | Yes | ||
| 37 | DNAJC11 | 1483 | 4.794 | 0.4491 | Yes | ||
| 38 | PCSK4 | 1730 | 4.220 | 0.4431 | Yes | ||
| 39 | PRIM1 | 1905 | 3.839 | 0.4403 | Yes | ||
| 40 | HNRPA1 | 1915 | 3.826 | 0.4465 | Yes | ||
| 41 | DNMT1 | 1929 | 3.795 | 0.4523 | Yes | ||
| 42 | MAT2A | 1931 | 3.792 | 0.4589 | Yes | ||
| 43 | APRIN | 1985 | 3.694 | 0.4624 | Yes | ||
| 44 | MELK | 1988 | 3.692 | 0.4687 | Yes | ||
| 45 | FUS | 2012 | 3.648 | 0.4738 | Yes | ||
| 46 | TIAM1 | 2353 | 3.081 | 0.4607 | Yes | ||
| 47 | HTF9C | 2494 | 2.917 | 0.4582 | Yes | ||
| 48 | CACNA1G | 2496 | 2.914 | 0.4632 | Yes | ||
| 49 | CENPB | 2558 | 2.829 | 0.4648 | Yes | ||
| 50 | SFRS1 | 2638 | 2.745 | 0.4653 | Yes | ||
| 51 | MYC | 2641 | 2.742 | 0.4699 | Yes | ||
| 52 | E2F3 | 2656 | 2.722 | 0.4739 | Yes | ||
| 53 | KLHDC3 | 2695 | 2.671 | 0.4765 | Yes | ||
| 54 | DLST | 2724 | 2.634 | 0.4795 | Yes | ||
| 55 | PAQR4 | 2953 | 2.433 | 0.4714 | No | ||
| 56 | UXT | 3220 | 2.204 | 0.4608 | No | ||
| 57 | SLBP | 3263 | 2.174 | 0.4623 | No | ||
| 58 | EIF2S1 | 3611 | 1.939 | 0.4469 | No | ||
| 59 | NR6A1 | 3687 | 1.892 | 0.4461 | No | ||
| 60 | PHF15 | 3703 | 1.882 | 0.4485 | No | ||
| 61 | STMN1 | 3958 | 1.757 | 0.4378 | No | ||
| 62 | ATAD2 | 4101 | 1.689 | 0.4330 | No | ||
| 63 | PODN | 4198 | 1.642 | 0.4307 | No | ||
| 64 | DDB2 | 4296 | 1.593 | 0.4282 | No | ||
| 65 | TREX2 | 4497 | 1.511 | 0.4200 | No | ||
| 66 | NR3C2 | 4645 | 1.454 | 0.4145 | No | ||
| 67 | PLK4 | 4862 | 1.371 | 0.4052 | No | ||
| 68 | E2F7 | 4863 | 1.370 | 0.4076 | No | ||
| 69 | KCTD15 | 4913 | 1.350 | 0.4073 | No | ||
| 70 | GRIA4 | 5109 | 1.285 | 0.3989 | No | ||
| 71 | SEZ6 | 5327 | 1.214 | 0.3893 | No | ||
| 72 | DAXX | 5361 | 1.199 | 0.3896 | No | ||
| 73 | EVI1 | 5872 | 1.043 | 0.3637 | No | ||
| 74 | RBBP4 | 6021 | 0.998 | 0.3575 | No | ||
| 75 | EPHB1 | 6336 | 0.901 | 0.3420 | No | ||
| 76 | PCSK1 | 6456 | 0.865 | 0.3371 | No | ||
| 77 | SEMA5A | 7228 | 0.664 | 0.2964 | No | ||
| 78 | SLITRK3 | 7415 | 0.613 | 0.2874 | No | ||
| 79 | SALL1 | 7519 | 0.582 | 0.2828 | No | ||
| 80 | FBXO5 | 7550 | 0.575 | 0.2822 | No | ||
| 81 | ASXL2 | 7563 | 0.573 | 0.2825 | No | ||
| 82 | PRKDC | 8026 | 0.454 | 0.2583 | No | ||
| 83 | ARHGEF17 | 8179 | 0.415 | 0.2508 | No | ||
| 84 | PDGFRA | 8201 | 0.410 | 0.2504 | No | ||
| 85 | HNRPD | 8252 | 0.397 | 0.2483 | No | ||
| 86 | HIST1H1D | 8291 | 0.389 | 0.2469 | No | ||
| 87 | LGALS1 | 8682 | 0.295 | 0.2263 | No | ||
| 88 | NUP153 | 9114 | 0.191 | 0.2033 | No | ||
| 89 | E2F1 | 9178 | 0.176 | 0.2002 | No | ||
| 90 | GLRA3 | 9286 | 0.147 | 0.1946 | No | ||
| 91 | RBL1 | 9387 | 0.123 | 0.1894 | No | ||
| 92 | TOPBP1 | 9684 | 0.052 | 0.1735 | No | ||
| 93 | UFD1L | 10061 | -0.048 | 0.1532 | No | ||
| 94 | ZIC3 | 10075 | -0.051 | 0.1526 | No | ||
| 95 | PLAGL1 | 10087 | -0.053 | 0.1521 | No | ||
| 96 | DMC1 | 10246 | -0.090 | 0.1437 | No | ||
| 97 | REPS2 | 10400 | -0.129 | 0.1356 | No | ||
| 98 | CASP8AP2 | 10586 | -0.177 | 0.1259 | No | ||
| 99 | JPH1 | 10669 | -0.200 | 0.1218 | No | ||
| 100 | PTMA | 10754 | -0.223 | 0.1176 | No | ||
| 101 | ADAMTS2 | 10773 | -0.227 | 0.1170 | No | ||
| 102 | H2AFZ | 10924 | -0.270 | 0.1094 | No | ||
| 103 | EIF4G2 | 10936 | -0.272 | 0.1092 | No | ||
| 104 | SF4 | 10990 | -0.284 | 0.1069 | No | ||
| 105 | HMGN2 | 11070 | -0.312 | 0.1031 | No | ||
| 106 | IPO7 | 11203 | -0.344 | 0.0966 | No | ||
| 107 | RHD | 11269 | -0.366 | 0.0937 | No | ||
| 108 | TBX3 | 11295 | -0.372 | 0.0930 | No | ||
| 109 | STAG1 | 11462 | -0.415 | 0.0847 | No | ||
| 110 | DNAJC9 | 11962 | -0.548 | 0.0586 | No | ||
| 111 | FMO4 | 11981 | -0.554 | 0.0586 | No | ||
| 112 | KLF5 | 12208 | -0.624 | 0.0474 | No | ||
| 113 | ZCCHC8 | 12347 | -0.664 | 0.0411 | No | ||
| 114 | DLL4 | 12405 | -0.682 | 0.0392 | No | ||
| 115 | AP4M1 | 12483 | -0.706 | 0.0362 | No | ||
| 116 | DMD | 12944 | -0.850 | 0.0128 | No | ||
| 117 | RPS20 | 12983 | -0.865 | 0.0122 | No | ||
| 118 | DCLRE1A | 13064 | -0.899 | 0.0094 | No | ||
| 119 | SLC16A2 | 13065 | -0.899 | 0.0110 | No | ||
| 120 | NRIP3 | 13089 | -0.907 | 0.0113 | No | ||
| 121 | DIO3 | 13174 | -0.938 | 0.0084 | No | ||
| 122 | CBX5 | 13421 | -1.026 | -0.0032 | No | ||
| 123 | TLE3 | 13477 | -1.055 | -0.0043 | No | ||
| 124 | RET | 13491 | -1.063 | -0.0032 | No | ||
| 125 | PEG3 | 13581 | -1.098 | -0.0061 | No | ||
| 126 | A2BP1 | 13618 | -1.110 | -0.0061 | No | ||
| 127 | CCNT2 | 13622 | -1.112 | -0.0043 | No | ||
| 128 | TOP1 | 14010 | -1.275 | -0.0231 | No | ||
| 129 | STAG2 | 14159 | -1.349 | -0.0288 | No | ||
| 130 | EGLN2 | 14181 | -1.357 | -0.0276 | No | ||
| 131 | FANCG | 14189 | -1.360 | -0.0256 | No | ||
| 132 | BRMS1L | 14296 | -1.417 | -0.0289 | No | ||
| 133 | RPL28 | 14398 | -1.472 | -0.0318 | No | ||
| 134 | ANP32E | 14730 | -1.653 | -0.0469 | No | ||
| 135 | ATP6V1D | 14746 | -1.659 | -0.0448 | No | ||
| 136 | VAMP3 | 14867 | -1.728 | -0.0483 | No | ||
| 137 | ATE1 | 14949 | -1.776 | -0.0496 | No | ||
| 138 | DPYSL2 | 15060 | -1.859 | -0.0523 | No | ||
| 139 | USP52 | 15078 | -1.869 | -0.0500 | No | ||
| 140 | ARF3 | 15160 | -1.919 | -0.0511 | No | ||
| 141 | LUC7L2 | 15248 | -1.975 | -0.0524 | No | ||
| 142 | ARID4A | 15277 | -1.998 | -0.0504 | No | ||
| 143 | BMP7 | 15459 | -2.168 | -0.0565 | No | ||
| 144 | RNF121 | 15722 | -2.428 | -0.0664 | No | ||
| 145 | PVRL1 | 15848 | -2.554 | -0.0688 | No | ||
| 146 | HNRPA0 | 15885 | -2.594 | -0.0662 | No | ||
| 147 | HIST1H2AH | 15886 | -2.595 | -0.0617 | No | ||
| 148 | ARHGAP6 | 15966 | -2.700 | -0.0613 | No | ||
| 149 | MAZ | 16104 | -2.868 | -0.0638 | No | ||
| 150 | FBXL20 | 16370 | -3.282 | -0.0724 | No | ||
| 151 | AXUD1 | 16377 | -3.295 | -0.0671 | No | ||
| 152 | NHLRC2 | 16553 | -3.677 | -0.0702 | No | ||
| 153 | CTDSP1 | 16610 | -3.781 | -0.0666 | No | ||
| 154 | HRBL | 16761 | -4.122 | -0.0676 | No | ||
| 155 | ING3 | 17233 | -5.277 | -0.0840 | No | ||
| 156 | PIM1 | 17322 | -5.526 | -0.0791 | No | ||
| 157 | CTCF | 17449 | -5.920 | -0.0757 | No | ||
| 158 | EZH2 | 17453 | -5.929 | -0.0656 | No | ||
| 159 | POLD1 | 17475 | -5.968 | -0.0564 | No | ||
| 160 | DCK | 17594 | -6.389 | -0.0517 | No | ||
| 161 | RPS6KA5 | 17680 | -6.679 | -0.0447 | No | ||
| 162 | SLC25A11 | 17732 | -6.850 | -0.0355 | No | ||
| 163 | PPM1D | 17752 | -6.926 | -0.0246 | No | ||
| 164 | CDKN1A | 17893 | -7.620 | -0.0189 | No | ||
| 165 | PPP1R9B | 17968 | -8.059 | -0.0089 | No | ||
| 166 | RAB11B | 18175 | -9.658 | -0.0033 | No | ||
| 167 | MXD3 | 18489 | -15.662 | 0.0069 | No |