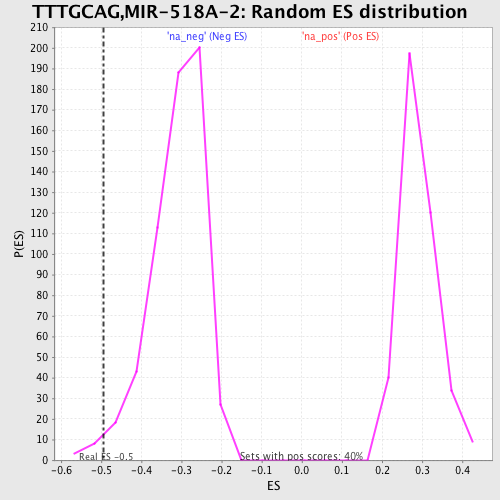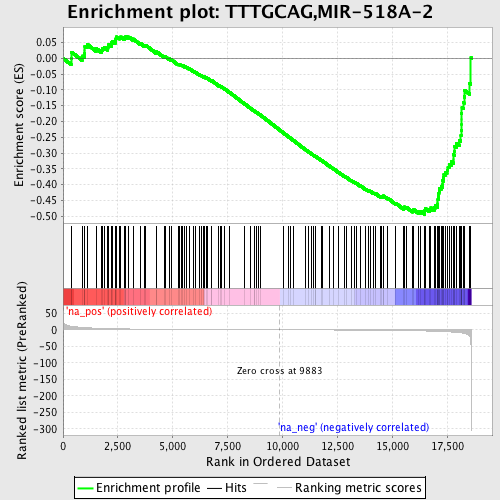
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTTGCAG,MIR-518A-2 |
| Enrichment Score (ES) | -0.49479613 |
| Normalized Enrichment Score (NES) | -1.5957603 |
| Nominal p-value | 0.018333333 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC25A26 | 367 | 10.547 | 0.0004 | No | ||
| 2 | ETF1 | 378 | 10.464 | 0.0200 | No | ||
| 3 | IVNS1ABP | 866 | 7.144 | 0.0074 | No | ||
| 4 | PPM1F | 970 | 6.589 | 0.0144 | No | ||
| 5 | NEDD8 | 992 | 6.500 | 0.0258 | No | ||
| 6 | RBBP7 | 993 | 6.495 | 0.0383 | No | ||
| 7 | PICALM | 1096 | 6.067 | 0.0444 | No | ||
| 8 | TARDBP | 1504 | 4.726 | 0.0315 | No | ||
| 9 | FUSIP1 | 1750 | 4.189 | 0.0263 | No | ||
| 10 | MMD | 1799 | 4.056 | 0.0315 | No | ||
| 11 | UBAP2 | 1875 | 3.892 | 0.0349 | No | ||
| 12 | RHOT1 | 2043 | 3.590 | 0.0328 | No | ||
| 13 | BCL2 | 2046 | 3.584 | 0.0395 | No | ||
| 14 | CDK2AP1 | 2084 | 3.519 | 0.0443 | No | ||
| 15 | HMGA1 | 2217 | 3.288 | 0.0435 | No | ||
| 16 | SPG20 | 2218 | 3.287 | 0.0498 | No | ||
| 17 | CORO1C | 2252 | 3.234 | 0.0542 | No | ||
| 18 | CTDP1 | 2369 | 3.046 | 0.0538 | No | ||
| 19 | SEH1L | 2391 | 3.027 | 0.0585 | No | ||
| 20 | EIF3S1 | 2409 | 3.006 | 0.0633 | No | ||
| 21 | SET | 2435 | 2.972 | 0.0677 | No | ||
| 22 | TOMM70A | 2559 | 2.829 | 0.0665 | No | ||
| 23 | EGR3 | 2636 | 2.748 | 0.0677 | No | ||
| 24 | PABPN1 | 2805 | 2.559 | 0.0635 | No | ||
| 25 | CUL2 | 2834 | 2.533 | 0.0668 | No | ||
| 26 | DHX15 | 2860 | 2.511 | 0.0703 | No | ||
| 27 | HMGB1 | 2996 | 2.390 | 0.0676 | No | ||
| 28 | SEPT6 | 3193 | 2.216 | 0.0613 | No | ||
| 29 | KPNA3 | 3532 | 1.989 | 0.0468 | No | ||
| 30 | QKI | 3701 | 1.887 | 0.0413 | No | ||
| 31 | CNN1 | 3758 | 1.853 | 0.0418 | No | ||
| 32 | ANKRD17 | 4249 | 1.611 | 0.0184 | No | ||
| 33 | ATP11A | 4252 | 1.610 | 0.0214 | No | ||
| 34 | ADRA2B | 4610 | 1.464 | 0.0049 | No | ||
| 35 | NR3C2 | 4645 | 1.454 | 0.0058 | No | ||
| 36 | NCKIPSD | 4830 | 1.384 | -0.0015 | No | ||
| 37 | DSCAML1 | 4919 | 1.348 | -0.0036 | No | ||
| 38 | DSC3 | 5245 | 1.242 | -0.0188 | No | ||
| 39 | ZIC1 | 5285 | 1.225 | -0.0186 | No | ||
| 40 | OSBPL8 | 5382 | 1.192 | -0.0215 | No | ||
| 41 | LEF1 | 5437 | 1.177 | -0.0222 | No | ||
| 42 | ACTR3 | 5544 | 1.145 | -0.0257 | No | ||
| 43 | BTBD11 | 5627 | 1.118 | -0.0280 | No | ||
| 44 | CATSPER2 | 5738 | 1.084 | -0.0319 | No | ||
| 45 | AEBP2 | 5930 | 1.027 | -0.0402 | No | ||
| 46 | HMGA2 | 6045 | 0.991 | -0.0445 | No | ||
| 47 | GDF11 | 6229 | 0.933 | -0.0526 | No | ||
| 48 | RAI2 | 6285 | 0.915 | -0.0538 | No | ||
| 49 | R3HDM1 | 6412 | 0.878 | -0.0590 | No | ||
| 50 | GUCY2C | 6424 | 0.875 | -0.0579 | No | ||
| 51 | AFF2 | 6515 | 0.849 | -0.0611 | No | ||
| 52 | PNN | 6588 | 0.828 | -0.0634 | No | ||
| 53 | DDX42 | 6769 | 0.782 | -0.0717 | No | ||
| 54 | LAMC1 | 6771 | 0.782 | -0.0702 | No | ||
| 55 | SGCZ | 7101 | 0.698 | -0.0867 | No | ||
| 56 | KPNA4 | 7150 | 0.686 | -0.0880 | No | ||
| 57 | TAF4 | 7198 | 0.674 | -0.0892 | No | ||
| 58 | DKK2 | 7357 | 0.628 | -0.0966 | No | ||
| 59 | SAMD4A | 7572 | 0.571 | -0.1071 | No | ||
| 60 | PRKAB2 | 8272 | 0.393 | -0.1441 | No | ||
| 61 | TMEPAI | 8552 | 0.323 | -0.1586 | No | ||
| 62 | POU4F1 | 8733 | 0.283 | -0.1678 | No | ||
| 63 | RASGRP4 | 8734 | 0.282 | -0.1673 | No | ||
| 64 | WDR26 | 8812 | 0.260 | -0.1710 | No | ||
| 65 | KCNJ2 | 8904 | 0.240 | -0.1754 | No | ||
| 66 | ACVR2A | 8986 | 0.218 | -0.1794 | No | ||
| 67 | MYOG | 10033 | -0.041 | -0.2359 | No | ||
| 68 | ANK2 | 10261 | -0.094 | -0.2480 | No | ||
| 69 | SP8 | 10345 | -0.115 | -0.2523 | No | ||
| 70 | PTGFRN | 10484 | -0.151 | -0.2595 | No | ||
| 71 | JAG1 | 11026 | -0.296 | -0.2882 | No | ||
| 72 | DAPK1 | 11174 | -0.339 | -0.2955 | No | ||
| 73 | NDRG1 | 11342 | -0.382 | -0.3038 | No | ||
| 74 | PAX3 | 11425 | -0.404 | -0.3075 | No | ||
| 75 | CREB3L2 | 11486 | -0.422 | -0.3099 | No | ||
| 76 | SIPA1L2 | 11508 | -0.428 | -0.3102 | No | ||
| 77 | ATRN | 11757 | -0.495 | -0.3227 | No | ||
| 78 | STK38 | 11799 | -0.506 | -0.3240 | No | ||
| 79 | HNRPR | 12118 | -0.592 | -0.3400 | No | ||
| 80 | AMACR | 12300 | -0.649 | -0.3486 | No | ||
| 81 | SATB2 | 12568 | -0.734 | -0.3616 | No | ||
| 82 | FOXA1 | 12811 | -0.812 | -0.3732 | No | ||
| 83 | PLCB1 | 12902 | -0.841 | -0.3764 | No | ||
| 84 | NCOR1 | 13128 | -0.926 | -0.3868 | No | ||
| 85 | CPEB2 | 13280 | -0.975 | -0.3931 | No | ||
| 86 | TMEM32 | 13356 | -1.002 | -0.3953 | No | ||
| 87 | CEPT1 | 13534 | -1.081 | -0.4028 | No | ||
| 88 | INHBB | 13800 | -1.182 | -0.4149 | No | ||
| 89 | SRCRB4D | 13927 | -1.234 | -0.4193 | No | ||
| 90 | PHF6 | 13999 | -1.270 | -0.4207 | No | ||
| 91 | STAG2 | 14159 | -1.349 | -0.4267 | No | ||
| 92 | PACSIN1 | 14236 | -1.385 | -0.4282 | No | ||
| 93 | PTPRF | 14475 | -1.517 | -0.4381 | No | ||
| 94 | THRAP2 | 14513 | -1.536 | -0.4372 | No | ||
| 95 | SRPK2 | 14580 | -1.572 | -0.4377 | No | ||
| 96 | PLEKHA3 | 14604 | -1.582 | -0.4359 | No | ||
| 97 | ARPP-19 | 14788 | -1.679 | -0.4426 | No | ||
| 98 | KCMF1 | 15163 | -1.924 | -0.4592 | No | ||
| 99 | EIF4G3 | 15532 | -2.244 | -0.4748 | No | ||
| 100 | TIMP2 | 15541 | -2.252 | -0.4709 | No | ||
| 101 | DYRK1A | 15661 | -2.363 | -0.4728 | No | ||
| 102 | FBXL5 | 15927 | -2.658 | -0.4820 | No | ||
| 103 | CGGBP1 | 15984 | -2.724 | -0.4798 | No | ||
| 104 | YTHDF3 | 16189 | -2.994 | -0.4851 | No | ||
| 105 | DOCK10 | 16297 | -3.179 | -0.4848 | No | ||
| 106 | GNB4 | 16483 | -3.509 | -0.4880 | Yes | ||
| 107 | CHD9 | 16488 | -3.525 | -0.4815 | Yes | ||
| 108 | SURF4 | 16500 | -3.559 | -0.4752 | Yes | ||
| 109 | MAPRE1 | 16691 | -3.930 | -0.4780 | Yes | ||
| 110 | CNNM2 | 16765 | -4.133 | -0.4740 | Yes | ||
| 111 | EIF2C2 | 16911 | -4.445 | -0.4733 | Yes | ||
| 112 | MEIS1 | 16952 | -4.525 | -0.4667 | Yes | ||
| 113 | PFN2 | 17060 | -4.747 | -0.4634 | Yes | ||
| 114 | RAP2C | 17082 | -4.821 | -0.4553 | Yes | ||
| 115 | SLC39A9 | 17083 | -4.828 | -0.4460 | Yes | ||
| 116 | RAB6A | 17108 | -4.921 | -0.4378 | Yes | ||
| 117 | RANBP10 | 17123 | -4.953 | -0.4291 | Yes | ||
| 118 | LAMP2 | 17134 | -4.980 | -0.4200 | Yes | ||
| 119 | NRF1 | 17165 | -5.084 | -0.4119 | Yes | ||
| 120 | RAB21 | 17229 | -5.266 | -0.4052 | Yes | ||
| 121 | RAB22A | 17298 | -5.476 | -0.3983 | Yes | ||
| 122 | PLEKHB2 | 17304 | -5.481 | -0.3881 | Yes | ||
| 123 | BIRC4 | 17326 | -5.532 | -0.3786 | Yes | ||
| 124 | BRI3 | 17357 | -5.658 | -0.3693 | Yes | ||
| 125 | RNF11 | 17412 | -5.803 | -0.3611 | Yes | ||
| 126 | HRB | 17498 | -6.026 | -0.3541 | Yes | ||
| 127 | PPP1R12A | 17541 | -6.198 | -0.3445 | Yes | ||
| 128 | PURB | 17595 | -6.397 | -0.3350 | Yes | ||
| 129 | BCL11A | 17700 | -6.753 | -0.3277 | Yes | ||
| 130 | ARRDC3 | 17795 | -7.118 | -0.3191 | Yes | ||
| 131 | MAP3K3 | 17796 | -7.121 | -0.3054 | Yes | ||
| 132 | CD47 | 17841 | -7.316 | -0.2937 | Yes | ||
| 133 | PJA2 | 17850 | -7.377 | -0.2799 | Yes | ||
| 134 | MKRN1 | 17918 | -7.743 | -0.2687 | Yes | ||
| 135 | NEDD4L | 18073 | -8.721 | -0.2603 | Yes | ||
| 136 | CDC42EP3 | 18131 | -9.250 | -0.2456 | Yes | ||
| 137 | IHPK1 | 18142 | -9.332 | -0.2282 | Yes | ||
| 138 | PLEKHM1 | 18155 | -9.426 | -0.2107 | Yes | ||
| 139 | DUSP6 | 18160 | -9.525 | -0.1926 | Yes | ||
| 140 | TCF12 | 18161 | -9.536 | -0.1743 | Yes | ||
| 141 | WDR22 | 18179 | -9.670 | -0.1566 | Yes | ||
| 142 | ITCH | 18253 | -10.384 | -0.1406 | Yes | ||
| 143 | ZFP36L1 | 18278 | -10.574 | -0.1215 | Yes | ||
| 144 | GADD45A | 18307 | -10.892 | -0.1021 | Yes | ||
| 145 | JARID1B | 18522 | -17.914 | -0.0793 | Yes | ||
| 146 | LRRN3 | 18589 | -43.847 | 0.0015 | Yes |