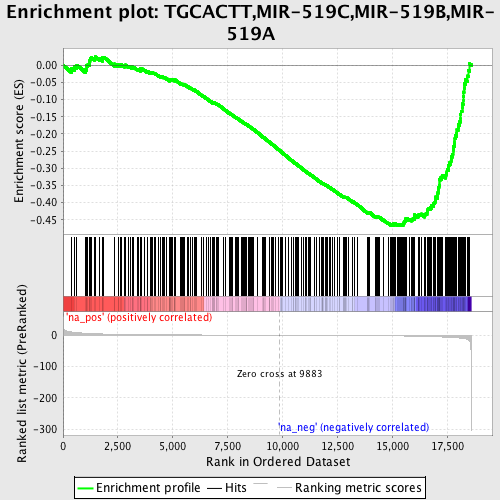
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TGCACTT,MIR-519C,MIR-519B,MIR-519A |
| Enrichment Score (ES) | -0.4679385 |
| Normalized Enrichment Score (NES) | -1.6195308 |
| Nominal p-value | 0.003322259 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NR1D1 | 387 | 10.397 | -0.0084 | No | ||
| 2 | TAF5L | 523 | 9.328 | -0.0044 | No | ||
| 3 | MCM7 | 624 | 8.576 | 0.0006 | No | ||
| 4 | EFTUD1 | 1019 | 6.389 | -0.0131 | No | ||
| 5 | NIPA1 | 1065 | 6.201 | -0.0079 | No | ||
| 6 | SMOC1 | 1070 | 6.174 | -0.0006 | No | ||
| 7 | ZFYVE9 | 1132 | 5.945 | 0.0033 | No | ||
| 8 | KCND2 | 1204 | 5.719 | 0.0065 | No | ||
| 9 | SUV39H1 | 1211 | 5.672 | 0.0131 | No | ||
| 10 | LRRIQ2 | 1226 | 5.627 | 0.0192 | No | ||
| 11 | YTHDF2 | 1277 | 5.489 | 0.0232 | No | ||
| 12 | SFRS2 | 1426 | 4.971 | 0.0212 | No | ||
| 13 | SFRS5 | 1470 | 4.834 | 0.0247 | No | ||
| 14 | AHCTF1 | 1660 | 4.369 | 0.0198 | No | ||
| 15 | CDK2 | 1797 | 4.060 | 0.0173 | No | ||
| 16 | KLF9 | 1798 | 4.058 | 0.0223 | No | ||
| 17 | KCNJ10 | 1856 | 3.931 | 0.0239 | No | ||
| 18 | MOBKL2B | 2340 | 3.102 | 0.0014 | No | ||
| 19 | ATP2C1 | 2360 | 3.059 | 0.0041 | No | ||
| 20 | HABP4 | 2503 | 2.908 | -0.0001 | No | ||
| 21 | MIB1 | 2542 | 2.852 | 0.0013 | No | ||
| 22 | BIRC6 | 2611 | 2.774 | 0.0010 | No | ||
| 23 | DNAJB6 | 2652 | 2.725 | 0.0021 | No | ||
| 24 | ST8SIA2 | 2784 | 2.571 | -0.0019 | No | ||
| 25 | FOXF2 | 2815 | 2.547 | -0.0004 | No | ||
| 26 | SNX16 | 2844 | 2.524 | 0.0011 | No | ||
| 27 | DDX3X | 2995 | 2.391 | -0.0041 | No | ||
| 28 | PCDHA12 | 3051 | 2.342 | -0.0043 | No | ||
| 29 | NEUROG2 | 3143 | 2.259 | -0.0065 | No | ||
| 30 | WEE1 | 3159 | 2.243 | -0.0046 | No | ||
| 31 | CELSR2 | 3221 | 2.203 | -0.0052 | No | ||
| 32 | WDFY3 | 3381 | 2.086 | -0.0113 | No | ||
| 33 | M6PR | 3420 | 2.058 | -0.0109 | No | ||
| 34 | KPNA3 | 3532 | 1.989 | -0.0145 | No | ||
| 35 | FASTK | 3537 | 1.987 | -0.0123 | No | ||
| 36 | SKI | 3538 | 1.986 | -0.0099 | No | ||
| 37 | PCDHA1 | 3550 | 1.979 | -0.0081 | No | ||
| 38 | QKI | 3701 | 1.887 | -0.0139 | No | ||
| 39 | MAP3K12 | 3851 | 1.808 | -0.0199 | No | ||
| 40 | KCNN2 | 3856 | 1.806 | -0.0179 | No | ||
| 41 | STYX | 3979 | 1.748 | -0.0224 | No | ||
| 42 | LRCH2 | 3996 | 1.741 | -0.0211 | No | ||
| 43 | CRSP2 | 4022 | 1.726 | -0.0204 | No | ||
| 44 | MYT1 | 4069 | 1.703 | -0.0208 | No | ||
| 45 | VLDLR | 4148 | 1.667 | -0.0230 | No | ||
| 46 | BHLHB3 | 4219 | 1.627 | -0.0249 | No | ||
| 47 | PKIA | 4358 | 1.563 | -0.0305 | No | ||
| 48 | RAB10 | 4438 | 1.534 | -0.0329 | No | ||
| 49 | MAP3K9 | 4442 | 1.532 | -0.0312 | No | ||
| 50 | PIGS | 4512 | 1.505 | -0.0332 | No | ||
| 51 | ZFPM2 | 4581 | 1.477 | -0.0351 | No | ||
| 52 | PTHLH | 4608 | 1.464 | -0.0347 | No | ||
| 53 | LIMK1 | 4712 | 1.424 | -0.0386 | No | ||
| 54 | PHTF2 | 4848 | 1.376 | -0.0443 | No | ||
| 55 | E2F7 | 4863 | 1.370 | -0.0434 | No | ||
| 56 | HOXB3 | 4870 | 1.367 | -0.0420 | No | ||
| 57 | HOXD8 | 4872 | 1.366 | -0.0404 | No | ||
| 58 | HAS2 | 4914 | 1.350 | -0.0410 | No | ||
| 59 | MAPRE3 | 4930 | 1.344 | -0.0402 | No | ||
| 60 | PARD6B | 4992 | 1.329 | -0.0419 | No | ||
| 61 | LRIG1 | 5065 | 1.298 | -0.0442 | No | ||
| 62 | NPAS2 | 5069 | 1.297 | -0.0428 | No | ||
| 63 | LAPTM4A | 5090 | 1.291 | -0.0423 | No | ||
| 64 | RYR3 | 5123 | 1.280 | -0.0425 | No | ||
| 65 | SFMBT1 | 5354 | 1.202 | -0.0536 | No | ||
| 66 | SPRY4 | 5410 | 1.183 | -0.0551 | No | ||
| 67 | KLF12 | 5423 | 1.180 | -0.0543 | No | ||
| 68 | SLC1A2 | 5465 | 1.170 | -0.0552 | No | ||
| 69 | HIF1A | 5533 | 1.149 | -0.0574 | No | ||
| 70 | KIF23 | 5551 | 1.142 | -0.0569 | No | ||
| 71 | KIF5A | 5661 | 1.107 | -0.0615 | No | ||
| 72 | SYN2 | 5707 | 1.092 | -0.0627 | No | ||
| 73 | DDX11 | 5792 | 1.064 | -0.0659 | No | ||
| 74 | ACVR1 | 5821 | 1.057 | -0.0662 | No | ||
| 75 | ERBB4 | 5918 | 1.029 | -0.0702 | No | ||
| 76 | MAPK6 | 5965 | 1.017 | -0.0714 | No | ||
| 77 | HMGA2 | 6045 | 0.991 | -0.0745 | No | ||
| 78 | CREB5 | 6076 | 0.981 | -0.0750 | No | ||
| 79 | NKIRAS1 | 6325 | 0.903 | -0.0874 | No | ||
| 80 | R3HDM1 | 6412 | 0.878 | -0.0910 | No | ||
| 81 | RKHD1 | 6517 | 0.849 | -0.0957 | No | ||
| 82 | POLQ | 6646 | 0.813 | -0.1017 | No | ||
| 83 | MAPK4 | 6726 | 0.792 | -0.1050 | No | ||
| 84 | PTPN2 | 6823 | 0.768 | -0.1093 | No | ||
| 85 | ANKRD29 | 6826 | 0.767 | -0.1085 | No | ||
| 86 | FBXW11 | 6840 | 0.764 | -0.1083 | No | ||
| 87 | NFIA | 6866 | 0.757 | -0.1087 | No | ||
| 88 | TNFRSF21 | 6890 | 0.753 | -0.1090 | No | ||
| 89 | SMOC2 | 6892 | 0.752 | -0.1082 | No | ||
| 90 | CASP2 | 7004 | 0.723 | -0.1134 | No | ||
| 91 | LMX1A | 7009 | 0.722 | -0.1127 | No | ||
| 92 | STC1 | 7040 | 0.714 | -0.1135 | No | ||
| 93 | ENPP5 | 7084 | 0.702 | -0.1150 | No | ||
| 94 | FJX1 | 7306 | 0.642 | -0.1262 | No | ||
| 95 | SLITRK3 | 7415 | 0.613 | -0.1314 | No | ||
| 96 | ASXL2 | 7563 | 0.573 | -0.1387 | No | ||
| 97 | SLC24A4 | 7608 | 0.563 | -0.1404 | No | ||
| 98 | PANX2 | 7655 | 0.550 | -0.1423 | No | ||
| 99 | IGF1 | 7738 | 0.530 | -0.1461 | No | ||
| 100 | WDR20 | 7739 | 0.530 | -0.1454 | No | ||
| 101 | PCDHA10 | 7868 | 0.492 | -0.1518 | No | ||
| 102 | VANGL1 | 7904 | 0.484 | -0.1532 | No | ||
| 103 | SMAD7 | 7954 | 0.471 | -0.1553 | No | ||
| 104 | ROCK2 | 7984 | 0.465 | -0.1563 | No | ||
| 105 | PCDHA3 | 8008 | 0.457 | -0.1570 | No | ||
| 106 | PCDHA11 | 8113 | 0.431 | -0.1621 | No | ||
| 107 | PCDH20 | 8155 | 0.420 | -0.1638 | No | ||
| 108 | PDGFRA | 8201 | 0.410 | -0.1658 | No | ||
| 109 | SYT1 | 8280 | 0.390 | -0.1696 | No | ||
| 110 | DDX3Y | 8305 | 0.385 | -0.1704 | No | ||
| 111 | RBBP6 | 8335 | 0.379 | -0.1715 | No | ||
| 112 | ZBTB4 | 8355 | 0.373 | -0.1721 | No | ||
| 113 | HIVEP2 | 8358 | 0.371 | -0.1718 | No | ||
| 114 | FSTL5 | 8471 | 0.343 | -0.1775 | No | ||
| 115 | RAPGEF4 | 8490 | 0.339 | -0.1781 | No | ||
| 116 | ZDHHC1 | 8538 | 0.327 | -0.1802 | No | ||
| 117 | EMX2 | 8607 | 0.311 | -0.1836 | No | ||
| 118 | NFIX | 8641 | 0.303 | -0.1850 | No | ||
| 119 | ABHD3 | 8660 | 0.300 | -0.1856 | No | ||
| 120 | KLF11 | 8670 | 0.298 | -0.1857 | No | ||
| 121 | PGM2L1 | 8870 | 0.249 | -0.1963 | No | ||
| 122 | TNFSF12 | 9109 | 0.192 | -0.2091 | No | ||
| 123 | TESK2 | 9115 | 0.191 | -0.2091 | No | ||
| 124 | BTBD7 | 9121 | 0.189 | -0.2091 | No | ||
| 125 | E2F1 | 9178 | 0.176 | -0.2120 | No | ||
| 126 | PKD2 | 9225 | 0.165 | -0.2143 | No | ||
| 127 | NBL1 | 9400 | 0.119 | -0.2236 | No | ||
| 128 | ARHGAP12 | 9416 | 0.115 | -0.2243 | No | ||
| 129 | NPAT | 9425 | 0.111 | -0.2246 | No | ||
| 130 | WNK3 | 9514 | 0.091 | -0.2293 | No | ||
| 131 | FAM60A | 9543 | 0.084 | -0.2307 | No | ||
| 132 | DCBLD2 | 9584 | 0.074 | -0.2328 | No | ||
| 133 | HEY2 | 9677 | 0.054 | -0.2378 | No | ||
| 134 | GDA | 9794 | 0.025 | -0.2441 | No | ||
| 135 | SASH1 | 9889 | -0.002 | -0.2492 | No | ||
| 136 | NHLH1 | 9935 | -0.015 | -0.2517 | No | ||
| 137 | SUMF1 | 9949 | -0.018 | -0.2524 | No | ||
| 138 | ZFHX4 | 9995 | -0.031 | -0.2548 | No | ||
| 139 | SLC40A1 | 10131 | -0.064 | -0.2621 | No | ||
| 140 | KIAA1715 | 10272 | -0.096 | -0.2696 | No | ||
| 141 | SP3 | 10431 | -0.137 | -0.2781 | No | ||
| 142 | PTGFRN | 10484 | -0.151 | -0.2807 | No | ||
| 143 | PRKAA1 | 10511 | -0.156 | -0.2820 | No | ||
| 144 | RTN2 | 10578 | -0.174 | -0.2853 | No | ||
| 145 | PCDHA7 | 10595 | -0.178 | -0.2860 | No | ||
| 146 | EPHA4 | 10624 | -0.186 | -0.2873 | No | ||
| 147 | TSG101 | 10699 | -0.209 | -0.2911 | No | ||
| 148 | EIF2C1 | 10720 | -0.215 | -0.2919 | No | ||
| 149 | SHANK2 | 10855 | -0.250 | -0.2989 | No | ||
| 150 | EIF4G2 | 10936 | -0.272 | -0.3030 | No | ||
| 151 | COL4A3 | 11042 | -0.301 | -0.3083 | No | ||
| 152 | MTMR4 | 11064 | -0.310 | -0.3091 | No | ||
| 153 | CACNB1 | 11109 | -0.323 | -0.3111 | No | ||
| 154 | CCNJ | 11196 | -0.343 | -0.3154 | No | ||
| 155 | TEAD1 | 11235 | -0.353 | -0.3170 | No | ||
| 156 | THRA | 11254 | -0.362 | -0.3176 | No | ||
| 157 | TBX3 | 11295 | -0.372 | -0.3193 | No | ||
| 158 | YES1 | 11451 | -0.412 | -0.3273 | No | ||
| 159 | ANUBL1 | 11459 | -0.415 | -0.3271 | No | ||
| 160 | SOCS6 | 11559 | -0.442 | -0.3320 | No | ||
| 161 | COL19A1 | 11689 | -0.477 | -0.3385 | No | ||
| 162 | ATRN | 11757 | -0.495 | -0.3415 | No | ||
| 163 | STK38 | 11799 | -0.506 | -0.3431 | No | ||
| 164 | PCDHA5 | 11839 | -0.515 | -0.3446 | No | ||
| 165 | RORB | 11844 | -0.516 | -0.3442 | No | ||
| 166 | NHLH2 | 11854 | -0.520 | -0.3441 | No | ||
| 167 | TOPORS | 11953 | -0.547 | -0.3488 | No | ||
| 168 | CALD1 | 11956 | -0.547 | -0.3482 | No | ||
| 169 | RASD1 | 12011 | -0.561 | -0.3505 | No | ||
| 170 | MAP3K5 | 12019 | -0.563 | -0.3502 | No | ||
| 171 | MASTL | 12064 | -0.576 | -0.3519 | No | ||
| 172 | SH3BP5 | 12131 | -0.598 | -0.3547 | No | ||
| 173 | DNAJB9 | 12176 | -0.614 | -0.3564 | No | ||
| 174 | EREG | 12297 | -0.649 | -0.3621 | No | ||
| 175 | NEUROG1 | 12351 | -0.666 | -0.3642 | No | ||
| 176 | PTPN4 | 12488 | -0.707 | -0.3708 | No | ||
| 177 | RTN1 | 12579 | -0.737 | -0.3748 | No | ||
| 178 | HOXA3 | 12604 | -0.746 | -0.3752 | No | ||
| 179 | BRWD1 | 12792 | -0.803 | -0.3844 | No | ||
| 180 | GRM7 | 12800 | -0.807 | -0.3838 | No | ||
| 181 | FOXA1 | 12811 | -0.812 | -0.3834 | No | ||
| 182 | GOLGA1 | 12843 | -0.822 | -0.3841 | No | ||
| 183 | SNX5 | 12856 | -0.826 | -0.3837 | No | ||
| 184 | PIK3R1 | 12882 | -0.833 | -0.3841 | No | ||
| 185 | PLCB1 | 12902 | -0.841 | -0.3841 | No | ||
| 186 | NRP2 | 13021 | -0.880 | -0.3894 | No | ||
| 187 | TBL1X | 13185 | -0.943 | -0.3972 | No | ||
| 188 | GOSR1 | 13204 | -0.950 | -0.3970 | No | ||
| 189 | CPEB2 | 13280 | -0.975 | -0.3999 | No | ||
| 190 | CNOT7 | 13416 | -1.022 | -0.4060 | No | ||
| 191 | TNFSF11 | 13876 | -1.210 | -0.4296 | No | ||
| 192 | MAB21L1 | 13879 | -1.211 | -0.4282 | No | ||
| 193 | BMPR2 | 13916 | -1.226 | -0.4287 | No | ||
| 194 | ATG16L1 | 13921 | -1.230 | -0.4274 | No | ||
| 195 | MAP3K8 | 13974 | -1.256 | -0.4287 | No | ||
| 196 | TRIM3 | 14219 | -1.376 | -0.4404 | No | ||
| 197 | HMGB3 | 14240 | -1.388 | -0.4398 | No | ||
| 198 | BRMS1L | 14296 | -1.417 | -0.4410 | No | ||
| 199 | FGF9 | 14329 | -1.436 | -0.4410 | No | ||
| 200 | DUSP8 | 14373 | -1.458 | -0.4416 | No | ||
| 201 | CSNK1G1 | 14418 | -1.481 | -0.4422 | No | ||
| 202 | PLEKHA3 | 14604 | -1.582 | -0.4504 | No | ||
| 203 | HIF1AN | 14818 | -1.696 | -0.4599 | No | ||
| 204 | SNIP1 | 14924 | -1.757 | -0.4635 | No | ||
| 205 | PPP3CA | 14970 | -1.793 | -0.4638 | No | ||
| 206 | BLCAP | 15008 | -1.824 | -0.4636 | No | ||
| 207 | TNRC6A | 15052 | -1.856 | -0.4636 | No | ||
| 208 | DPYSL2 | 15060 | -1.859 | -0.4617 | No | ||
| 209 | PCDHA6 | 15082 | -1.871 | -0.4606 | No | ||
| 210 | KCNQ2 | 15138 | -1.906 | -0.4613 | No | ||
| 211 | PAPOLA | 15251 | -1.976 | -0.4650 | No | ||
| 212 | ARID4A | 15277 | -1.998 | -0.4639 | No | ||
| 213 | PTPRT | 15340 | -2.054 | -0.4648 | No | ||
| 214 | ITPR1 | 15399 | -2.111 | -0.4654 | Yes | ||
| 215 | SOX21 | 15407 | -2.121 | -0.4631 | Yes | ||
| 216 | BCL2L2 | 15457 | -2.165 | -0.4632 | Yes | ||
| 217 | BICD2 | 15497 | -2.200 | -0.4626 | Yes | ||
| 218 | EFNB1 | 15500 | -2.206 | -0.4600 | Yes | ||
| 219 | BNC2 | 15533 | -2.244 | -0.4590 | Yes | ||
| 220 | TIMP2 | 15541 | -2.252 | -0.4567 | Yes | ||
| 221 | UBE3A | 15569 | -2.269 | -0.4554 | Yes | ||
| 222 | ARHGAP1 | 15585 | -2.289 | -0.4534 | Yes | ||
| 223 | L3MBTL3 | 15610 | -2.312 | -0.4519 | Yes | ||
| 224 | CRKRS | 15617 | -2.315 | -0.4494 | Yes | ||
| 225 | PAFAH1B1 | 15623 | -2.326 | -0.4468 | Yes | ||
| 226 | DYRK1A | 15661 | -2.363 | -0.4459 | Yes | ||
| 227 | ARHGEF11 | 15798 | -2.493 | -0.4503 | Yes | ||
| 228 | NF1 | 15859 | -2.566 | -0.4504 | Yes | ||
| 229 | MAP3K2 | 15889 | -2.600 | -0.4488 | Yes | ||
| 230 | FBXL5 | 15927 | -2.658 | -0.4476 | Yes | ||
| 231 | RAD52 | 15981 | -2.722 | -0.4472 | Yes | ||
| 232 | DDX5 | 16002 | -2.745 | -0.4449 | Yes | ||
| 233 | STX6 | 16009 | -2.751 | -0.4419 | Yes | ||
| 234 | OGT | 16017 | -2.763 | -0.4389 | Yes | ||
| 235 | NFAT5 | 16018 | -2.763 | -0.4355 | Yes | ||
| 236 | CFL2 | 16176 | -2.956 | -0.4404 | Yes | ||
| 237 | LACE1 | 16194 | -2.998 | -0.4377 | Yes | ||
| 238 | TNKS1BP1 | 16216 | -3.033 | -0.4351 | Yes | ||
| 239 | CUGBP2 | 16257 | -3.116 | -0.4335 | Yes | ||
| 240 | ACTL6A | 16320 | -3.211 | -0.4330 | Yes | ||
| 241 | PIGA | 16485 | -3.515 | -0.4376 | Yes | ||
| 242 | CHD9 | 16488 | -3.525 | -0.4334 | Yes | ||
| 243 | RB1 | 16538 | -3.642 | -0.4317 | Yes | ||
| 244 | USP15 | 16587 | -3.735 | -0.4297 | Yes | ||
| 245 | RNF31 | 16591 | -3.743 | -0.4253 | Yes | ||
| 246 | MMP14 | 16601 | -3.759 | -0.4212 | Yes | ||
| 247 | MYB | 16630 | -3.818 | -0.4180 | Yes | ||
| 248 | MAPRE1 | 16691 | -3.930 | -0.4165 | Yes | ||
| 249 | SERTAD2 | 16751 | -4.090 | -0.4147 | Yes | ||
| 250 | SNRK | 16785 | -4.176 | -0.4114 | Yes | ||
| 251 | GNS | 16803 | -4.207 | -0.4072 | Yes | ||
| 252 | SMAD5 | 16886 | -4.371 | -0.4063 | Yes | ||
| 253 | ZHX2 | 16890 | -4.381 | -0.4012 | Yes | ||
| 254 | ADIPOR2 | 16941 | -4.498 | -0.3984 | Yes | ||
| 255 | ERBB2IP | 16954 | -4.527 | -0.3935 | Yes | ||
| 256 | CUL3 | 16971 | -4.550 | -0.3888 | Yes | ||
| 257 | DAZAP2 | 16976 | -4.558 | -0.3835 | Yes | ||
| 258 | PFN2 | 17060 | -4.747 | -0.3822 | Yes | ||
| 259 | GOPC | 17070 | -4.783 | -0.3768 | Yes | ||
| 260 | RAP2C | 17082 | -4.821 | -0.3715 | Yes | ||
| 261 | MECP2 | 17092 | -4.861 | -0.3661 | Yes | ||
| 262 | NDEL1 | 17096 | -4.875 | -0.3603 | Yes | ||
| 263 | RFX3 | 17120 | -4.942 | -0.3555 | Yes | ||
| 264 | LAMP2 | 17134 | -4.980 | -0.3501 | Yes | ||
| 265 | SLC4A7 | 17157 | -5.060 | -0.3451 | Yes | ||
| 266 | GTPBP2 | 17158 | -5.060 | -0.3389 | Yes | ||
| 267 | ZDHHC9 | 17159 | -5.063 | -0.3327 | Yes | ||
| 268 | USP3 | 17187 | -5.127 | -0.3279 | Yes | ||
| 269 | SEMA4B | 17259 | -5.357 | -0.3253 | Yes | ||
| 270 | NAGK | 17283 | -5.447 | -0.3199 | Yes | ||
| 271 | WDR1 | 17441 | -5.897 | -0.3212 | Yes | ||
| 272 | AKT3 | 17454 | -5.931 | -0.3146 | Yes | ||
| 273 | ACSL4 | 17494 | -6.013 | -0.3094 | Yes | ||
| 274 | DNM2 | 17496 | -6.025 | -0.3021 | Yes | ||
| 275 | HECA | 17565 | -6.292 | -0.2981 | Yes | ||
| 276 | ARID4B | 17570 | -6.303 | -0.2906 | Yes | ||
| 277 | PURB | 17595 | -6.397 | -0.2841 | Yes | ||
| 278 | PPP1R10 | 17678 | -6.674 | -0.2804 | Yes | ||
| 279 | ABHD2 | 17684 | -6.692 | -0.2725 | Yes | ||
| 280 | PIK4CB | 17701 | -6.755 | -0.2651 | Yes | ||
| 281 | MBNL1 | 17740 | -6.866 | -0.2588 | Yes | ||
| 282 | SORL1 | 17771 | -7.017 | -0.2518 | Yes | ||
| 283 | TGFBR2 | 17774 | -7.023 | -0.2434 | Yes | ||
| 284 | MAP3K3 | 17796 | -7.121 | -0.2358 | Yes | ||
| 285 | EPC2 | 17827 | -7.228 | -0.2286 | Yes | ||
| 286 | KBTBD2 | 17854 | -7.427 | -0.2209 | Yes | ||
| 287 | ASF1A | 17858 | -7.457 | -0.2120 | Yes | ||
| 288 | SSH2 | 17889 | -7.595 | -0.2043 | Yes | ||
| 289 | CNOT4 | 17933 | -7.853 | -0.1971 | Yes | ||
| 290 | MAML1 | 17950 | -7.943 | -0.1882 | Yes | ||
| 291 | TRIOBP | 18003 | -8.295 | -0.1809 | Yes | ||
| 292 | STK17B | 18027 | -8.461 | -0.1718 | Yes | ||
| 293 | PKNOX1 | 18082 | -8.811 | -0.1640 | Yes | ||
| 294 | CIC | 18106 | -9.023 | -0.1542 | Yes | ||
| 295 | SCAMP2 | 18112 | -9.061 | -0.1434 | Yes | ||
| 296 | ELK3 | 18134 | -9.258 | -0.1332 | Yes | ||
| 297 | STAT3 | 18184 | -9.748 | -0.1240 | Yes | ||
| 298 | BTG3 | 18191 | -9.860 | -0.1123 | Yes | ||
| 299 | RAB5B | 18231 | -10.238 | -0.1019 | Yes | ||
| 300 | VAMP4 | 18237 | -10.317 | -0.0895 | Yes | ||
| 301 | ITCH | 18253 | -10.384 | -0.0776 | Yes | ||
| 302 | SOX4 | 18273 | -10.557 | -0.0658 | Yes | ||
| 303 | ANKRD50 | 18286 | -10.628 | -0.0534 | Yes | ||
| 304 | ANKFY1 | 18328 | -11.197 | -0.0420 | Yes | ||
| 305 | ARHGAP24 | 18437 | -13.832 | -0.0309 | Yes | ||
| 306 | HBP1 | 18472 | -15.082 | -0.0144 | Yes | ||
| 307 | SOX5 | 18526 | -18.118 | 0.0049 | Yes |
