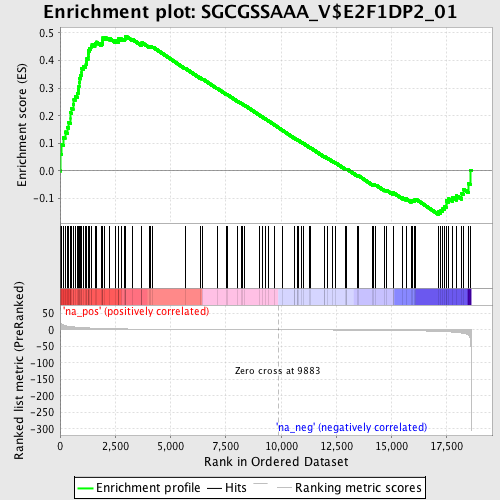
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | SGCGSSAAA_V$E2F1DP2_01 |
| Enrichment Score (ES) | 0.4886331 |
| Normalized Enrichment Score (NES) | 1.6718205 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03691412 |
| FWER p-Value | 0.408 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MCM6 | 9 | 30.323 | 0.0613 | Yes | ||
| 2 | RANBP1 | 55 | 18.855 | 0.0973 | Yes | ||
| 3 | AK2 | 153 | 13.651 | 0.1199 | Yes | ||
| 4 | SNRPD1 | 224 | 12.313 | 0.1412 | Yes | ||
| 5 | UNG | 340 | 10.863 | 0.1571 | Yes | ||
| 6 | ACBD6 | 382 | 10.443 | 0.1762 | Yes | ||
| 7 | RQCD1 | 462 | 9.734 | 0.1917 | Yes | ||
| 8 | PHF5A | 464 | 9.691 | 0.2114 | Yes | ||
| 9 | CDC6 | 518 | 9.366 | 0.2277 | Yes | ||
| 10 | CDCA7 | 587 | 8.803 | 0.2419 | Yes | ||
| 11 | MCM7 | 624 | 8.576 | 0.2575 | Yes | ||
| 12 | SYNCRIP | 679 | 8.203 | 0.2713 | Yes | ||
| 13 | MCM3 | 785 | 7.561 | 0.2810 | Yes | ||
| 14 | NCL | 817 | 7.411 | 0.2944 | Yes | ||
| 15 | MRPL40 | 848 | 7.236 | 0.3075 | Yes | ||
| 16 | NOLC1 | 859 | 7.172 | 0.3216 | Yes | ||
| 17 | FKBP5 | 889 | 6.987 | 0.3343 | Yes | ||
| 18 | MSH2 | 911 | 6.898 | 0.3472 | Yes | ||
| 19 | CDC25A | 963 | 6.668 | 0.3580 | Yes | ||
| 20 | NASP | 969 | 6.607 | 0.3712 | Yes | ||
| 21 | MCM4 | 1059 | 6.251 | 0.3792 | Yes | ||
| 22 | AP1S1 | 1170 | 5.839 | 0.3851 | Yes | ||
| 23 | KCNA6 | 1188 | 5.799 | 0.3960 | Yes | ||
| 24 | SUV39H1 | 1211 | 5.672 | 0.4064 | Yes | ||
| 25 | MCM2 | 1266 | 5.511 | 0.4147 | Yes | ||
| 26 | GMNN | 1272 | 5.493 | 0.4256 | Yes | ||
| 27 | SMAD6 | 1293 | 5.438 | 0.4356 | Yes | ||
| 28 | CLSPN | 1322 | 5.334 | 0.4450 | Yes | ||
| 29 | PRPS1 | 1398 | 5.067 | 0.4512 | Yes | ||
| 30 | USP37 | 1442 | 4.939 | 0.4590 | Yes | ||
| 31 | ACO2 | 1580 | 4.561 | 0.4609 | Yes | ||
| 32 | FANCC | 1629 | 4.432 | 0.4673 | Yes | ||
| 33 | POLR2A | 1865 | 3.916 | 0.4626 | Yes | ||
| 34 | POLD3 | 1927 | 3.807 | 0.4671 | Yes | ||
| 35 | DNMT1 | 1929 | 3.795 | 0.4747 | Yes | ||
| 36 | HIRA | 1938 | 3.779 | 0.4820 | Yes | ||
| 37 | NUP62 | 2028 | 3.620 | 0.4846 | Yes | ||
| 38 | HMGA1 | 2217 | 3.288 | 0.4811 | Yes | ||
| 39 | HTF9C | 2494 | 2.917 | 0.4721 | Yes | ||
| 40 | SFRS1 | 2638 | 2.745 | 0.4700 | Yes | ||
| 41 | MYC | 2641 | 2.742 | 0.4755 | Yes | ||
| 42 | E2F3 | 2656 | 2.722 | 0.4803 | Yes | ||
| 43 | EED | 2787 | 2.570 | 0.4785 | Yes | ||
| 44 | PCNA | 2909 | 2.465 | 0.4770 | Yes | ||
| 45 | YBX2 | 2941 | 2.443 | 0.4803 | Yes | ||
| 46 | PAQR4 | 2953 | 2.433 | 0.4846 | Yes | ||
| 47 | ILF3 | 2971 | 2.413 | 0.4886 | Yes | ||
| 48 | GATA1 | 3289 | 2.153 | 0.4759 | No | ||
| 49 | HOXC10 | 3673 | 1.899 | 0.4591 | No | ||
| 50 | NR6A1 | 3687 | 1.892 | 0.4622 | No | ||
| 51 | PHF15 | 3703 | 1.882 | 0.4652 | No | ||
| 52 | SFRS10 | 4036 | 1.720 | 0.4508 | No | ||
| 53 | ATAD2 | 4101 | 1.689 | 0.4508 | No | ||
| 54 | PODN | 4198 | 1.642 | 0.4489 | No | ||
| 55 | SYNGR4 | 5652 | 1.109 | 0.3727 | No | ||
| 56 | SFRS7 | 6339 | 0.900 | 0.3374 | No | ||
| 57 | PCSK1 | 6456 | 0.865 | 0.3329 | No | ||
| 58 | TYRO3 | 7108 | 0.696 | 0.2991 | No | ||
| 59 | FBXO5 | 7550 | 0.575 | 0.2765 | No | ||
| 60 | ASXL2 | 7563 | 0.573 | 0.2770 | No | ||
| 61 | PRKDC | 8026 | 0.454 | 0.2530 | No | ||
| 62 | STK35 | 8211 | 0.409 | 0.2438 | No | ||
| 63 | HNRPD | 8252 | 0.397 | 0.2425 | No | ||
| 64 | ZBTB4 | 8355 | 0.373 | 0.2377 | No | ||
| 65 | MYH10 | 9019 | 0.212 | 0.2023 | No | ||
| 66 | E2F1 | 9178 | 0.176 | 0.1942 | No | ||
| 67 | GLRA3 | 9286 | 0.147 | 0.1887 | No | ||
| 68 | PHC1 | 9449 | 0.104 | 0.1801 | No | ||
| 69 | TOPBP1 | 9684 | 0.052 | 0.1676 | No | ||
| 70 | PLAGL1 | 10087 | -0.053 | 0.1460 | No | ||
| 71 | CASP8AP2 | 10586 | -0.177 | 0.1194 | No | ||
| 72 | GPRC5B | 10600 | -0.178 | 0.1191 | No | ||
| 73 | PTMA | 10754 | -0.223 | 0.1113 | No | ||
| 74 | NRK | 10766 | -0.225 | 0.1111 | No | ||
| 75 | ADAMTS2 | 10773 | -0.227 | 0.1113 | No | ||
| 76 | H2AFZ | 10924 | -0.270 | 0.1037 | No | ||
| 77 | PRPF4B | 11006 | -0.290 | 0.0999 | No | ||
| 78 | POLE2 | 11285 | -0.370 | 0.0856 | No | ||
| 79 | GABRB3 | 11349 | -0.384 | 0.0830 | No | ||
| 80 | DNAJC9 | 11962 | -0.548 | 0.0511 | No | ||
| 81 | FMO4 | 11981 | -0.554 | 0.0512 | No | ||
| 82 | HNRPR | 12118 | -0.592 | 0.0451 | No | ||
| 83 | ZCCHC8 | 12347 | -0.664 | 0.0341 | No | ||
| 84 | AP4M1 | 12483 | -0.706 | 0.0282 | No | ||
| 85 | UGCGL1 | 12933 | -0.847 | 0.0057 | No | ||
| 86 | ATF5 | 12975 | -0.863 | 0.0052 | No | ||
| 87 | RPS20 | 12983 | -0.865 | 0.0066 | No | ||
| 88 | KCNS2 | 13458 | -1.044 | -0.0169 | No | ||
| 89 | RET | 13491 | -1.063 | -0.0164 | No | ||
| 90 | STAG2 | 14159 | -1.349 | -0.0497 | No | ||
| 91 | FANCG | 14189 | -1.360 | -0.0485 | No | ||
| 92 | BRMS1L | 14296 | -1.417 | -0.0514 | No | ||
| 93 | FHOD1 | 14694 | -1.634 | -0.0695 | No | ||
| 94 | EFNA5 | 14763 | -1.670 | -0.0698 | No | ||
| 95 | USP52 | 15078 | -1.869 | -0.0829 | No | ||
| 96 | HNRPUL1 | 15081 | -1.869 | -0.0792 | No | ||
| 97 | TRIM39 | 15504 | -2.209 | -0.0975 | No | ||
| 98 | HCN3 | 15656 | -2.360 | -0.1009 | No | ||
| 99 | HIST1H2AH | 15886 | -2.595 | -0.1080 | No | ||
| 100 | ARHGAP6 | 15966 | -2.700 | -0.1067 | No | ||
| 101 | POLE4 | 16044 | -2.798 | -0.1052 | No | ||
| 102 | MAZ | 16104 | -2.868 | -0.1025 | No | ||
| 103 | ALDH6A1 | 17130 | -4.971 | -0.1478 | No | ||
| 104 | ING3 | 17233 | -5.277 | -0.1426 | No | ||
| 105 | PIM1 | 17322 | -5.526 | -0.1361 | No | ||
| 106 | RRM2 | 17403 | -5.780 | -0.1286 | No | ||
| 107 | POLD1 | 17475 | -5.968 | -0.1203 | No | ||
| 108 | HIST1H2BK | 17476 | -5.969 | -0.1081 | No | ||
| 109 | DCK | 17594 | -6.389 | -0.1014 | No | ||
| 110 | PPM1D | 17752 | -6.926 | -0.0958 | No | ||
| 111 | HIST1H4A | 17939 | -7.890 | -0.0898 | No | ||
| 112 | MSH5 | 18170 | -9.624 | -0.0826 | No | ||
| 113 | H2AFV | 18254 | -10.388 | -0.0659 | No | ||
| 114 | MXD3 | 18489 | -15.662 | -0.0466 | No | ||
| 115 | IL4I1 | 18563 | -26.226 | 0.0029 | No |