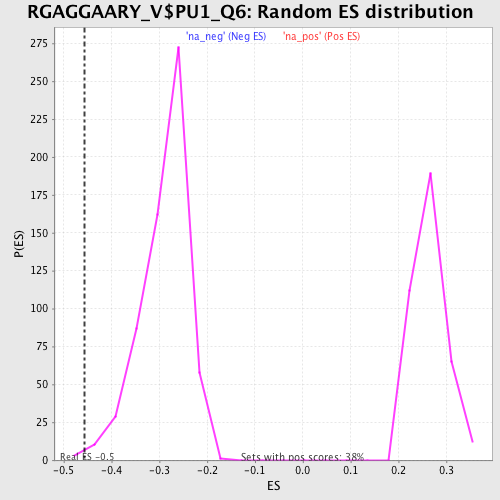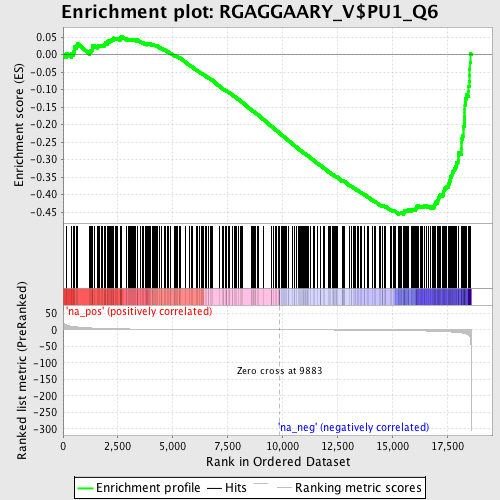
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RGAGGAARY_V$PU1_Q6 |
| Enrichment Score (ES) | -0.4565208 |
| Normalized Enrichment Score (NES) | -1.5846651 |
| Nominal p-value | 0.004823151 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | G6PC3 | 166 | 13.392 | 0.0039 | No | ||
| 2 | ETF1 | 378 | 10.464 | 0.0025 | No | ||
| 3 | ATPAF2 | 456 | 9.783 | 0.0077 | No | ||
| 4 | PPRC1 | 502 | 9.457 | 0.0144 | No | ||
| 5 | CDK8 | 527 | 9.306 | 0.0221 | No | ||
| 6 | TRIP13 | 627 | 8.553 | 0.0249 | No | ||
| 7 | SMS | 639 | 8.477 | 0.0325 | No | ||
| 8 | HPRT1 | 1215 | 5.665 | 0.0066 | No | ||
| 9 | ANP32A | 1241 | 5.582 | 0.0106 | No | ||
| 10 | GALE | 1300 | 5.417 | 0.0127 | No | ||
| 11 | SLC16A6 | 1340 | 5.273 | 0.0156 | No | ||
| 12 | LUC7L | 1349 | 5.250 | 0.0203 | No | ||
| 13 | YWHAZ | 1354 | 5.245 | 0.0251 | No | ||
| 14 | ELMO1 | 1427 | 4.971 | 0.0260 | No | ||
| 15 | PSIP1 | 1561 | 4.602 | 0.0232 | No | ||
| 16 | PABPC1 | 1592 | 4.530 | 0.0259 | No | ||
| 17 | PIK3R3 | 1649 | 4.388 | 0.0271 | No | ||
| 18 | CENTD2 | 1732 | 4.215 | 0.0267 | No | ||
| 19 | CKB | 1811 | 4.036 | 0.0263 | No | ||
| 20 | BAG1 | 1866 | 3.911 | 0.0271 | No | ||
| 21 | SEMA6B | 1889 | 3.868 | 0.0297 | No | ||
| 22 | POLD3 | 1927 | 3.807 | 0.0313 | No | ||
| 23 | SSBP4 | 1930 | 3.793 | 0.0349 | No | ||
| 24 | CENTA1 | 2021 | 3.635 | 0.0335 | No | ||
| 25 | RASIP1 | 2023 | 3.631 | 0.0369 | No | ||
| 26 | BCL2 | 2046 | 3.584 | 0.0392 | No | ||
| 27 | VASP | 2129 | 3.447 | 0.0381 | No | ||
| 28 | HOXC4 | 2148 | 3.411 | 0.0404 | No | ||
| 29 | STC2 | 2187 | 3.347 | 0.0415 | No | ||
| 30 | CSF1 | 2216 | 3.289 | 0.0432 | No | ||
| 31 | CORO1C | 2252 | 3.234 | 0.0444 | No | ||
| 32 | ADD3 | 2306 | 3.150 | 0.0445 | No | ||
| 33 | ELOVL6 | 2308 | 3.149 | 0.0475 | No | ||
| 34 | STARD13 | 2384 | 3.037 | 0.0464 | No | ||
| 35 | YWHAQ | 2419 | 2.993 | 0.0474 | No | ||
| 36 | CACNA1G | 2496 | 2.914 | 0.0461 | No | ||
| 37 | IL17RE | 2595 | 2.787 | 0.0434 | No | ||
| 38 | CUEDC1 | 2603 | 2.779 | 0.0457 | No | ||
| 39 | ICA1 | 2612 | 2.774 | 0.0479 | No | ||
| 40 | POFUT1 | 2644 | 2.737 | 0.0489 | No | ||
| 41 | E2F3 | 2656 | 2.722 | 0.0509 | No | ||
| 42 | PCDH10 | 2670 | 2.703 | 0.0528 | No | ||
| 43 | SLC39A14 | 2893 | 2.479 | 0.0431 | No | ||
| 44 | SCG3 | 2902 | 2.473 | 0.0450 | No | ||
| 45 | DLAT | 2992 | 2.395 | 0.0425 | No | ||
| 46 | SRP72 | 3020 | 2.369 | 0.0433 | No | ||
| 47 | PPP1R14C | 3075 | 2.310 | 0.0426 | No | ||
| 48 | MATN3 | 3096 | 2.295 | 0.0437 | No | ||
| 49 | PRICKLE1 | 3160 | 2.243 | 0.0424 | No | ||
| 50 | UXT | 3220 | 2.204 | 0.0413 | No | ||
| 51 | ADAM15 | 3258 | 2.177 | 0.0414 | No | ||
| 52 | PTGS1 | 3284 | 2.157 | 0.0421 | No | ||
| 53 | WDFY3 | 3381 | 2.086 | 0.0389 | No | ||
| 54 | TIMELESS | 3384 | 2.085 | 0.0408 | No | ||
| 55 | KCNQ1 | 3387 | 2.082 | 0.0427 | No | ||
| 56 | PTPN7 | 3545 | 1.982 | 0.0360 | No | ||
| 57 | TFB2M | 3603 | 1.944 | 0.0348 | No | ||
| 58 | ITGB7 | 3653 | 1.915 | 0.0340 | No | ||
| 59 | NTS | 3757 | 1.853 | 0.0301 | No | ||
| 60 | CDC5L | 3807 | 1.831 | 0.0292 | No | ||
| 61 | DHX30 | 3816 | 1.829 | 0.0306 | No | ||
| 62 | CLSTN3 | 3829 | 1.820 | 0.0317 | No | ||
| 63 | AR | 3846 | 1.814 | 0.0325 | No | ||
| 64 | ANGPT2 | 3871 | 1.796 | 0.0330 | No | ||
| 65 | PLA2G4A | 3922 | 1.774 | 0.0319 | No | ||
| 66 | SRC | 3975 | 1.750 | 0.0308 | No | ||
| 67 | HMGN3 | 4052 | 1.715 | 0.0283 | No | ||
| 68 | ATP5H | 4084 | 1.697 | 0.0282 | No | ||
| 69 | MLLT10 | 4107 | 1.686 | 0.0287 | No | ||
| 70 | SYT7 | 4174 | 1.654 | 0.0267 | No | ||
| 71 | ELN | 4213 | 1.633 | 0.0262 | No | ||
| 72 | VDAC2 | 4236 | 1.619 | 0.0265 | No | ||
| 73 | NAV1 | 4282 | 1.597 | 0.0256 | No | ||
| 74 | SLC7A1 | 4408 | 1.546 | 0.0203 | No | ||
| 75 | TWIST1 | 4489 | 1.514 | 0.0174 | No | ||
| 76 | TREX2 | 4497 | 1.511 | 0.0184 | No | ||
| 77 | PTHLH | 4608 | 1.464 | 0.0138 | No | ||
| 78 | NR3C2 | 4645 | 1.454 | 0.0133 | No | ||
| 79 | SMOX | 4762 | 1.406 | 0.0083 | No | ||
| 80 | GPR85 | 4793 | 1.398 | 0.0080 | No | ||
| 81 | RHOJ | 4901 | 1.355 | 0.0035 | No | ||
| 82 | EFNA1 | 4915 | 1.349 | 0.0040 | No | ||
| 83 | CTBP2 | 5089 | 1.291 | -0.0042 | No | ||
| 84 | MAGED1 | 5093 | 1.291 | -0.0031 | No | ||
| 85 | WNT6 | 5139 | 1.274 | -0.0043 | No | ||
| 86 | SPRED2 | 5163 | 1.267 | -0.0043 | No | ||
| 87 | HR | 5193 | 1.257 | -0.0047 | No | ||
| 88 | PTPN22 | 5289 | 1.224 | -0.0087 | No | ||
| 89 | CYR61 | 5325 | 1.214 | -0.0095 | No | ||
| 90 | PTK7 | 5366 | 1.197 | -0.0105 | No | ||
| 91 | PLAT | 5585 | 1.133 | -0.0213 | No | ||
| 92 | CAPN3 | 5780 | 1.069 | -0.0309 | No | ||
| 93 | PAX8 | 5833 | 1.051 | -0.0327 | No | ||
| 94 | DHH | 5875 | 1.042 | -0.0340 | No | ||
| 95 | CREB5 | 6076 | 0.981 | -0.0440 | No | ||
| 96 | FLNC | 6130 | 0.965 | -0.0459 | No | ||
| 97 | HTR2B | 6233 | 0.931 | -0.0506 | No | ||
| 98 | TBC1D8 | 6287 | 0.915 | -0.0526 | No | ||
| 99 | DLC1 | 6333 | 0.902 | -0.0542 | No | ||
| 100 | TPM2 | 6406 | 0.880 | -0.0573 | No | ||
| 101 | ADCY4 | 6491 | 0.857 | -0.0611 | No | ||
| 102 | PCSK5 | 6554 | 0.836 | -0.0636 | No | ||
| 103 | IMPDH1 | 6608 | 0.822 | -0.0657 | No | ||
| 104 | ANGPTL1 | 6645 | 0.813 | -0.0669 | No | ||
| 105 | CACNA1D | 6718 | 0.794 | -0.0701 | No | ||
| 106 | HOXA1 | 6762 | 0.784 | -0.0717 | No | ||
| 107 | WNT9B | 6788 | 0.776 | -0.0723 | No | ||
| 108 | GCK | 7105 | 0.697 | -0.0889 | No | ||
| 109 | PRDM5 | 7278 | 0.650 | -0.0977 | No | ||
| 110 | ITGA11 | 7314 | 0.638 | -0.0990 | No | ||
| 111 | LGALS4 | 7323 | 0.636 | -0.0988 | No | ||
| 112 | NLGN3 | 7401 | 0.616 | -0.1024 | No | ||
| 113 | CDX2 | 7424 | 0.610 | -0.1030 | No | ||
| 114 | MMP9 | 7453 | 0.603 | -0.1040 | No | ||
| 115 | RAB2B | 7468 | 0.598 | -0.1042 | No | ||
| 116 | DMP1 | 7544 | 0.577 | -0.1077 | No | ||
| 117 | CA4 | 7588 | 0.567 | -0.1095 | No | ||
| 118 | KIRREL3 | 7602 | 0.565 | -0.1097 | No | ||
| 119 | C1QTNF5 | 7710 | 0.537 | -0.1150 | No | ||
| 120 | ACVRL1 | 7713 | 0.537 | -0.1146 | No | ||
| 121 | AHR | 7816 | 0.507 | -0.1197 | No | ||
| 122 | CD5 | 7876 | 0.490 | -0.1225 | No | ||
| 123 | MIST | 7913 | 0.483 | -0.1240 | No | ||
| 124 | RORA | 7973 | 0.468 | -0.1267 | No | ||
| 125 | CXXC5 | 8063 | 0.443 | -0.1312 | No | ||
| 126 | OTP | 8126 | 0.427 | -0.1341 | No | ||
| 127 | PCDHGA5 | 8188 | 0.412 | -0.1371 | No | ||
| 128 | RORC | 8567 | 0.320 | -0.1574 | No | ||
| 129 | EMX2 | 8607 | 0.311 | -0.1593 | No | ||
| 130 | NKX2-2 | 8634 | 0.305 | -0.1604 | No | ||
| 131 | RBMS3 | 8676 | 0.297 | -0.1624 | No | ||
| 132 | MTX2 | 8687 | 0.293 | -0.1626 | No | ||
| 133 | TNFSF13 | 8735 | 0.282 | -0.1649 | No | ||
| 134 | MSX1 | 8774 | 0.270 | -0.1667 | No | ||
| 135 | PAX6 | 8778 | 0.269 | -0.1666 | No | ||
| 136 | BMP5 | 8852 | 0.252 | -0.1704 | No | ||
| 137 | GPR44 | 8905 | 0.239 | -0.1730 | No | ||
| 138 | IL13 | 9135 | 0.186 | -0.1854 | No | ||
| 139 | CAMK2B | 9475 | 0.097 | -0.2038 | No | ||
| 140 | NINJ2 | 9482 | 0.096 | -0.2040 | No | ||
| 141 | EMCN | 9492 | 0.094 | -0.2044 | No | ||
| 142 | SEC14L2 | 9610 | 0.069 | -0.2108 | No | ||
| 143 | GNGT2 | 9678 | 0.054 | -0.2144 | No | ||
| 144 | PTGIR | 9710 | 0.047 | -0.2160 | No | ||
| 145 | PLCD1 | 9799 | 0.024 | -0.2208 | No | ||
| 146 | LTB4R | 9849 | 0.010 | -0.2235 | No | ||
| 147 | GPR56 | 9854 | 0.008 | -0.2237 | No | ||
| 148 | DOK2 | 9866 | 0.004 | -0.2243 | No | ||
| 149 | STAB1 | 9869 | 0.003 | -0.2244 | No | ||
| 150 | TMC3 | 9884 | -0.000 | -0.2252 | No | ||
| 151 | TLL1 | 9967 | -0.024 | -0.2296 | No | ||
| 152 | ETV6 | 10015 | -0.037 | -0.2322 | No | ||
| 153 | SH3TC1 | 10038 | -0.043 | -0.2333 | No | ||
| 154 | APOB48R | 10109 | -0.058 | -0.2371 | No | ||
| 155 | NDN | 10156 | -0.070 | -0.2396 | No | ||
| 156 | LTC4S | 10172 | -0.072 | -0.2403 | No | ||
| 157 | FGF20 | 10257 | -0.092 | -0.2448 | No | ||
| 158 | CXCR3 | 10280 | -0.097 | -0.2459 | No | ||
| 159 | SNCG | 10451 | -0.142 | -0.2551 | No | ||
| 160 | VIP | 10547 | -0.166 | -0.2601 | No | ||
| 161 | IL12B | 10559 | -0.169 | -0.2606 | No | ||
| 162 | TMPO | 10614 | -0.182 | -0.2633 | No | ||
| 163 | LYN | 10706 | -0.211 | -0.2681 | No | ||
| 164 | ADORA3 | 10757 | -0.223 | -0.2706 | No | ||
| 165 | HYAL3 | 10815 | -0.238 | -0.2735 | No | ||
| 166 | RERG | 10847 | -0.247 | -0.2750 | No | ||
| 167 | PLXNC1 | 10905 | -0.262 | -0.2778 | No | ||
| 168 | S100A10 | 10958 | -0.277 | -0.2804 | No | ||
| 169 | LEMD1 | 10976 | -0.280 | -0.2811 | No | ||
| 170 | PLCB2 | 10982 | -0.283 | -0.2811 | No | ||
| 171 | NDFIP1 | 11024 | -0.295 | -0.2830 | No | ||
| 172 | MMRN2 | 11036 | -0.298 | -0.2833 | No | ||
| 173 | IL17C | 11100 | -0.321 | -0.2865 | No | ||
| 174 | EVI2B | 11139 | -0.328 | -0.2882 | No | ||
| 175 | CNNM1 | 11142 | -0.329 | -0.2880 | No | ||
| 176 | SLC41A1 | 11206 | -0.346 | -0.2911 | No | ||
| 177 | ZFP161 | 11258 | -0.364 | -0.2936 | No | ||
| 178 | PPP1CC | 11390 | -0.396 | -0.3004 | No | ||
| 179 | PCDH9 | 11444 | -0.408 | -0.3029 | No | ||
| 180 | MARK3 | 11598 | -0.456 | -0.3108 | No | ||
| 181 | LCP2 | 11709 | -0.483 | -0.3163 | No | ||
| 182 | ESM1 | 11714 | -0.484 | -0.3161 | No | ||
| 183 | CSF3 | 11725 | -0.488 | -0.3162 | No | ||
| 184 | PITX2 | 11869 | -0.524 | -0.3235 | No | ||
| 185 | STAT5A | 11913 | -0.538 | -0.3253 | No | ||
| 186 | RAB39 | 12081 | -0.582 | -0.3339 | No | ||
| 187 | PBX4 | 12135 | -0.599 | -0.3362 | No | ||
| 188 | SLC12A4 | 12177 | -0.614 | -0.3379 | No | ||
| 189 | KIFC3 | 12276 | -0.643 | -0.3426 | No | ||
| 190 | CMYA5 | 12301 | -0.649 | -0.3433 | No | ||
| 191 | NCAM1 | 12337 | -0.661 | -0.3445 | No | ||
| 192 | EFNB3 | 12360 | -0.670 | -0.3451 | No | ||
| 193 | TYROBP | 12407 | -0.682 | -0.3470 | No | ||
| 194 | PLVAP | 12441 | -0.694 | -0.3481 | No | ||
| 195 | MAP2K1 | 12474 | -0.704 | -0.3492 | No | ||
| 196 | FGF14 | 12487 | -0.707 | -0.3491 | No | ||
| 197 | WNT3 | 12731 | -0.787 | -0.3617 | No | ||
| 198 | WNT5A | 12742 | -0.789 | -0.3615 | No | ||
| 199 | HOXC6 | 12748 | -0.791 | -0.3610 | No | ||
| 200 | GRIN2B | 12769 | -0.795 | -0.3613 | No | ||
| 201 | RAB20 | 12770 | -0.796 | -0.3605 | No | ||
| 202 | FOXA1 | 12811 | -0.812 | -0.3619 | No | ||
| 203 | SYTL1 | 12840 | -0.822 | -0.3627 | No | ||
| 204 | MPO | 13060 | -0.898 | -0.3738 | No | ||
| 205 | MAP4K2 | 13131 | -0.927 | -0.3767 | No | ||
| 206 | PTPRH | 13152 | -0.933 | -0.3769 | No | ||
| 207 | ITGAL | 13227 | -0.958 | -0.3800 | No | ||
| 208 | RNF12 | 13262 | -0.969 | -0.3809 | No | ||
| 209 | SOST | 13306 | -0.984 | -0.3823 | No | ||
| 210 | PLAGL2 | 13402 | -1.019 | -0.3865 | No | ||
| 211 | DNMT3A | 13484 | -1.060 | -0.3899 | No | ||
| 212 | MLLT7 | 13571 | -1.095 | -0.3936 | No | ||
| 213 | CSF2 | 13610 | -1.107 | -0.3946 | No | ||
| 214 | ITGA2B | 13731 | -1.147 | -0.4001 | No | ||
| 215 | TNF | 13737 | -1.150 | -0.3992 | No | ||
| 216 | TNFSF11 | 13876 | -1.210 | -0.4056 | No | ||
| 217 | FAM3D | 13918 | -1.227 | -0.4066 | No | ||
| 218 | SPATA6 | 14096 | -1.317 | -0.4151 | No | ||
| 219 | EGLN2 | 14181 | -1.357 | -0.4183 | No | ||
| 220 | ALDOA | 14258 | -1.395 | -0.4211 | No | ||
| 221 | RPL28 | 14398 | -1.472 | -0.4273 | No | ||
| 222 | MOBKL2C | 14471 | -1.515 | -0.4298 | No | ||
| 223 | EVX1 | 14542 | -1.550 | -0.4321 | No | ||
| 224 | DLX4 | 14550 | -1.552 | -0.4310 | No | ||
| 225 | TNK1 | 14560 | -1.559 | -0.4300 | No | ||
| 226 | TLR4 | 14638 | -1.600 | -0.4327 | No | ||
| 227 | LTA | 14700 | -1.636 | -0.4344 | No | ||
| 228 | LIF | 14708 | -1.641 | -0.4332 | No | ||
| 229 | REST | 14931 | -1.764 | -0.4436 | No | ||
| 230 | RALY | 14966 | -1.790 | -0.4438 | No | ||
| 231 | ERBB3 | 15038 | -1.849 | -0.4459 | No | ||
| 232 | HNRPUL1 | 15081 | -1.869 | -0.4464 | No | ||
| 233 | HOXB8 | 15136 | -1.904 | -0.4475 | No | ||
| 234 | HTR3B | 15291 | -2.015 | -0.4539 | No | ||
| 235 | MAX | 15307 | -2.029 | -0.4528 | No | ||
| 236 | DPP8 | 15350 | -2.064 | -0.4531 | No | ||
| 237 | FYN | 15378 | -2.088 | -0.4526 | No | ||
| 238 | ADORA1 | 15417 | -2.136 | -0.4526 | No | ||
| 239 | MR1 | 15420 | -2.138 | -0.4506 | No | ||
| 240 | RAB5C | 15529 | -2.242 | -0.4544 | Yes | ||
| 241 | BNC2 | 15533 | -2.244 | -0.4523 | Yes | ||
| 242 | RBM16 | 15537 | -2.248 | -0.4503 | Yes | ||
| 243 | TLN1 | 15543 | -2.253 | -0.4484 | Yes | ||
| 244 | SEPT1 | 15568 | -2.268 | -0.4475 | Yes | ||
| 245 | ATP1B3 | 15573 | -2.278 | -0.4456 | Yes | ||
| 246 | TNFSF14 | 15622 | -2.325 | -0.4459 | Yes | ||
| 247 | ORC4L | 15649 | -2.357 | -0.4451 | Yes | ||
| 248 | RAPGEF1 | 15702 | -2.405 | -0.4456 | Yes | ||
| 249 | CCR7 | 15734 | -2.436 | -0.4449 | Yes | ||
| 250 | RASGRP3 | 15746 | -2.446 | -0.4432 | Yes | ||
| 251 | LYL1 | 15758 | -2.462 | -0.4414 | Yes | ||
| 252 | RBM6 | 15893 | -2.606 | -0.4462 | Yes | ||
| 253 | RALA | 15896 | -2.609 | -0.4438 | Yes | ||
| 254 | SH2D3C | 15912 | -2.632 | -0.4421 | Yes | ||
| 255 | TCIRG1 | 15977 | -2.718 | -0.4429 | Yes | ||
| 256 | NFAT5 | 16018 | -2.763 | -0.4425 | Yes | ||
| 257 | ROCK1 | 16056 | -2.810 | -0.4418 | Yes | ||
| 258 | ANGPT1 | 16076 | -2.836 | -0.4401 | Yes | ||
| 259 | SRRM1 | 16093 | -2.852 | -0.4382 | Yes | ||
| 260 | CLIC1 | 16096 | -2.856 | -0.4355 | Yes | ||
| 261 | HAPLN1 | 16101 | -2.862 | -0.4330 | Yes | ||
| 262 | NAT6 | 16136 | -2.902 | -0.4320 | Yes | ||
| 263 | ITPR2 | 16186 | -2.985 | -0.4318 | Yes | ||
| 264 | ANGPTL2 | 16285 | -3.161 | -0.4341 | Yes | ||
| 265 | GALNT4 | 16324 | -3.218 | -0.4331 | Yes | ||
| 266 | FBXL20 | 16370 | -3.282 | -0.4324 | Yes | ||
| 267 | TPM3 | 16450 | -3.455 | -0.4334 | Yes | ||
| 268 | DMTF1 | 16467 | -3.487 | -0.4309 | Yes | ||
| 269 | NFYA | 16546 | -3.657 | -0.4316 | Yes | ||
| 270 | RGS12 | 16654 | -3.861 | -0.4337 | Yes | ||
| 271 | PIK3AP1 | 16744 | -4.064 | -0.4346 | Yes | ||
| 272 | CHD2 | 16826 | -4.251 | -0.4350 | Yes | ||
| 273 | ASB2 | 16891 | -4.382 | -0.4342 | Yes | ||
| 274 | CNOT6L | 16917 | -4.454 | -0.4313 | Yes | ||
| 275 | LRMP | 16940 | -4.496 | -0.4281 | Yes | ||
| 276 | ROD1 | 16958 | -4.531 | -0.4247 | Yes | ||
| 277 | MXD4 | 16968 | -4.544 | -0.4208 | Yes | ||
| 278 | SIPA1 | 17049 | -4.712 | -0.4206 | Yes | ||
| 279 | ZNF289 | 17077 | -4.803 | -0.4174 | Yes | ||
| 280 | VIM | 17089 | -4.844 | -0.4134 | Yes | ||
| 281 | PSEN1 | 17090 | -4.855 | -0.4087 | Yes | ||
| 282 | LAMP2 | 17134 | -4.980 | -0.4062 | Yes | ||
| 283 | TMOD3 | 17148 | -5.023 | -0.4020 | Yes | ||
| 284 | RUFY1 | 17183 | -5.124 | -0.3989 | Yes | ||
| 285 | FMNL1 | 17287 | -5.462 | -0.3993 | Yes | ||
| 286 | BIRC4 | 17326 | -5.532 | -0.3960 | Yes | ||
| 287 | IRAK4 | 17330 | -5.552 | -0.3908 | Yes | ||
| 288 | CREB3 | 17360 | -5.670 | -0.3869 | Yes | ||
| 289 | SLC9A3R1 | 17393 | -5.759 | -0.3831 | Yes | ||
| 290 | MTPN | 17413 | -5.809 | -0.3785 | Yes | ||
| 291 | ACSL4 | 17494 | -6.013 | -0.3771 | Yes | ||
| 292 | INPP5F | 17553 | -6.259 | -0.3742 | Yes | ||
| 293 | POU2AF1 | 17559 | -6.274 | -0.3684 | Yes | ||
| 294 | P2RY10 | 17601 | -6.419 | -0.3644 | Yes | ||
| 295 | ABI1 | 17609 | -6.457 | -0.3586 | Yes | ||
| 296 | ZFYVE1 | 17657 | -6.615 | -0.3548 | Yes | ||
| 297 | PTPRJ | 17664 | -6.637 | -0.3487 | Yes | ||
| 298 | PIK4CB | 17701 | -6.755 | -0.3441 | Yes | ||
| 299 | SLC1A5 | 17726 | -6.824 | -0.3388 | Yes | ||
| 300 | FGD2 | 17738 | -6.861 | -0.3328 | Yes | ||
| 301 | SPI1 | 17810 | -7.163 | -0.3297 | Yes | ||
| 302 | CD37 | 17851 | -7.406 | -0.3248 | Yes | ||
| 303 | ESRRA | 17890 | -7.595 | -0.3195 | Yes | ||
| 304 | TOB2 | 17921 | -7.756 | -0.3136 | Yes | ||
| 305 | PSCD4 | 17935 | -7.857 | -0.3067 | Yes | ||
| 306 | ICAM2 | 17999 | -8.264 | -0.3022 | Yes | ||
| 307 | PLD3 | 18015 | -8.386 | -0.2949 | Yes | ||
| 308 | CAMK1D | 18016 | -8.393 | -0.2868 | Yes | ||
| 309 | ASB7 | 18036 | -8.492 | -0.2796 | Yes | ||
| 310 | TCF12 | 18161 | -9.536 | -0.2772 | Yes | ||
| 311 | GIT2 | 18162 | -9.579 | -0.2679 | Yes | ||
| 312 | ABI3 | 18169 | -9.623 | -0.2589 | Yes | ||
| 313 | MSH5 | 18170 | -9.624 | -0.2496 | Yes | ||
| 314 | CD79B | 18176 | -9.666 | -0.2406 | Yes | ||
| 315 | TBCC | 18197 | -9.886 | -0.2321 | Yes | ||
| 316 | TRPV2 | 18232 | -10.268 | -0.2240 | Yes | ||
| 317 | PML | 18233 | -10.280 | -0.2141 | Yes | ||
| 318 | ORMDL3 | 18236 | -10.314 | -0.2042 | Yes | ||
| 319 | EIF4EBP1 | 18280 | -10.587 | -0.1963 | Yes | ||
| 320 | HHEX | 18291 | -10.672 | -0.1865 | Yes | ||
| 321 | VAV1 | 18300 | -10.774 | -0.1766 | Yes | ||
| 322 | POLD4 | 18311 | -10.982 | -0.1665 | Yes | ||
| 323 | AP1GBP1 | 18313 | -10.997 | -0.1559 | Yes | ||
| 324 | PTPRCAP | 18316 | -11.027 | -0.1454 | Yes | ||
| 325 | VGLL4 | 18325 | -11.131 | -0.1350 | Yes | ||
| 326 | PIK3CG | 18341 | -11.438 | -0.1248 | Yes | ||
| 327 | SAMSN1 | 18381 | -12.404 | -0.1149 | Yes | ||
| 328 | WASF2 | 18473 | -15.094 | -0.1053 | Yes | ||
| 329 | RAC2 | 18482 | -15.379 | -0.0909 | Yes | ||
| 330 | PTPN6 | 18504 | -16.488 | -0.0761 | Yes | ||
| 331 | SOX5 | 18526 | -18.118 | -0.0597 | Yes | ||
| 332 | TAX1BP3 | 18530 | -18.676 | -0.0418 | Yes | ||
| 333 | PDLIM2 | 18545 | -21.742 | -0.0215 | Yes | ||
| 334 | IL4I1 | 18563 | -26.226 | 0.0029 | Yes |