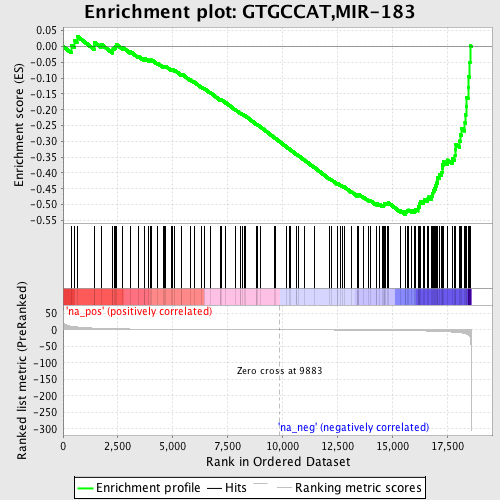
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GTGCCAT,MIR-183 |
| Enrichment Score (ES) | -0.531188 |
| Normalized Enrichment Score (NES) | -1.6586286 |
| Nominal p-value | 0.014571949 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VDAC1 | 377 | 10.480 | 0.0044 | No | ||
| 2 | PPP1R14B | 515 | 9.381 | 0.0191 | No | ||
| 3 | PPP2CB | 635 | 8.517 | 0.0328 | No | ||
| 4 | DTNA | 1413 | 5.021 | 0.0026 | No | ||
| 5 | SFRS2 | 1426 | 4.971 | 0.0137 | No | ||
| 6 | SOX6 | 1729 | 4.220 | 0.0073 | No | ||
| 7 | RNF138 | 2250 | 3.241 | -0.0132 | No | ||
| 8 | AMD1 | 2272 | 3.208 | -0.0067 | No | ||
| 9 | PKP4 | 2335 | 3.105 | -0.0028 | No | ||
| 10 | KCNK2 | 2402 | 3.014 | 0.0008 | No | ||
| 11 | ITGB1 | 2439 | 2.968 | 0.0058 | No | ||
| 12 | PDCD6 | 2710 | 2.646 | -0.0025 | No | ||
| 13 | FREQ | 3067 | 2.319 | -0.0163 | No | ||
| 14 | LPHN1 | 3416 | 2.060 | -0.0302 | No | ||
| 15 | QKI | 3701 | 1.887 | -0.0411 | No | ||
| 16 | PHF15 | 3703 | 1.882 | -0.0368 | No | ||
| 17 | IDH2 | 3890 | 1.789 | -0.0426 | No | ||
| 18 | SIN3A | 4001 | 1.739 | -0.0444 | No | ||
| 19 | GREM2 | 4040 | 1.720 | -0.0424 | No | ||
| 20 | TUB | 4293 | 1.594 | -0.0523 | No | ||
| 21 | ZFPM2 | 4581 | 1.477 | -0.0643 | No | ||
| 22 | KCNJ14 | 4598 | 1.468 | -0.0617 | No | ||
| 23 | EML4 | 4682 | 1.437 | -0.0628 | No | ||
| 24 | STK38L | 4940 | 1.341 | -0.0736 | No | ||
| 25 | LRP6 | 4965 | 1.336 | -0.0717 | No | ||
| 26 | LAPTM4A | 5090 | 1.291 | -0.0754 | No | ||
| 27 | OSBPL8 | 5382 | 1.192 | -0.0883 | No | ||
| 28 | MTMR6 | 5416 | 1.181 | -0.0873 | No | ||
| 29 | PLEKHA5 | 5799 | 1.062 | -0.1054 | No | ||
| 30 | SLC35A1 | 5970 | 1.016 | -0.1122 | No | ||
| 31 | ICK | 6312 | 0.909 | -0.1285 | No | ||
| 32 | BZW1 | 6427 | 0.874 | -0.1326 | No | ||
| 33 | RAI14 | 6699 | 0.798 | -0.1454 | No | ||
| 34 | PALM2 | 7151 | 0.686 | -0.1682 | No | ||
| 35 | TPM1 | 7164 | 0.683 | -0.1672 | No | ||
| 36 | HBEGF | 7211 | 0.671 | -0.1681 | No | ||
| 37 | SLITRK3 | 7415 | 0.613 | -0.1777 | No | ||
| 38 | HECTD2 | 7865 | 0.492 | -0.2008 | No | ||
| 39 | PEX19 | 8093 | 0.434 | -0.2120 | No | ||
| 40 | PAM | 8094 | 0.434 | -0.2110 | No | ||
| 41 | MAL2 | 8172 | 0.417 | -0.2142 | No | ||
| 42 | CDK5R1 | 8277 | 0.392 | -0.2189 | No | ||
| 43 | BRUNOL6 | 8299 | 0.387 | -0.2191 | No | ||
| 44 | CELSR3 | 8816 | 0.260 | -0.2464 | No | ||
| 45 | RCN2 | 8819 | 0.259 | -0.2459 | No | ||
| 46 | SEMA6D | 8844 | 0.254 | -0.2466 | No | ||
| 47 | ACVR2A | 8986 | 0.218 | -0.2537 | No | ||
| 48 | PLCB4 | 9613 | 0.069 | -0.2874 | No | ||
| 49 | UNC13B | 9658 | 0.060 | -0.2896 | No | ||
| 50 | RAB11FIP4 | 10187 | -0.075 | -0.3180 | No | ||
| 51 | TXNDC4 | 10301 | -0.102 | -0.3239 | No | ||
| 52 | IRS1 | 10369 | -0.122 | -0.3272 | No | ||
| 53 | TMPO | 10614 | -0.182 | -0.3400 | No | ||
| 54 | EPHA4 | 10624 | -0.186 | -0.3401 | No | ||
| 55 | TMSB4X | 10728 | -0.217 | -0.3451 | No | ||
| 56 | MAP3K4 | 11014 | -0.291 | -0.3598 | No | ||
| 57 | YES1 | 11451 | -0.412 | -0.3824 | No | ||
| 58 | ABCA1 | 12124 | -0.596 | -0.4174 | No | ||
| 59 | CHRD | 12252 | -0.635 | -0.4228 | No | ||
| 60 | PTPN4 | 12488 | -0.707 | -0.4338 | No | ||
| 61 | ARHGAP21 | 12495 | -0.710 | -0.4324 | No | ||
| 62 | DCX | 12632 | -0.756 | -0.4380 | No | ||
| 63 | PTDSS1 | 12734 | -0.788 | -0.4416 | No | ||
| 64 | PPP2R2A | 12805 | -0.809 | -0.4435 | No | ||
| 65 | VIL2 | 13149 | -0.932 | -0.4598 | No | ||
| 66 | PLAGL2 | 13402 | -1.019 | -0.4711 | No | ||
| 67 | PHLDB2 | 13432 | -1.031 | -0.4702 | No | ||
| 68 | AP3M1 | 13460 | -1.046 | -0.4692 | No | ||
| 69 | PDCD4 | 13480 | -1.055 | -0.4677 | No | ||
| 70 | CTDSPL | 13692 | -1.131 | -0.4765 | No | ||
| 71 | PLAG1 | 13919 | -1.229 | -0.4858 | No | ||
| 72 | PHF6 | 13999 | -1.270 | -0.4871 | No | ||
| 73 | SNX1 | 14266 | -1.401 | -0.4981 | No | ||
| 74 | BRMS1L | 14296 | -1.417 | -0.4964 | No | ||
| 75 | CSNK1G1 | 14418 | -1.481 | -0.4994 | No | ||
| 76 | SMPD3 | 14558 | -1.557 | -0.5033 | No | ||
| 77 | PLEKHA3 | 14604 | -1.582 | -0.5020 | No | ||
| 78 | ZDHHC6 | 14627 | -1.594 | -0.4994 | No | ||
| 79 | RALGDS | 14657 | -1.616 | -0.4971 | No | ||
| 80 | ANKRD13C | 14701 | -1.637 | -0.4956 | No | ||
| 81 | ARPP-19 | 14788 | -1.679 | -0.4963 | No | ||
| 82 | SNCB | 14827 | -1.702 | -0.4943 | No | ||
| 83 | PPP2CA | 15384 | -2.096 | -0.5195 | No | ||
| 84 | NCK2 | 15602 | -2.301 | -0.5258 | Yes | ||
| 85 | L3MBTL3 | 15610 | -2.312 | -0.5207 | Yes | ||
| 86 | BACH2 | 15673 | -2.376 | -0.5184 | Yes | ||
| 87 | PDE4D | 15754 | -2.452 | -0.5170 | Yes | ||
| 88 | RALA | 15896 | -2.609 | -0.5184 | Yes | ||
| 89 | OGT | 16017 | -2.763 | -0.5184 | Yes | ||
| 90 | CLCN3 | 16067 | -2.827 | -0.5144 | Yes | ||
| 91 | CFL2 | 16176 | -2.956 | -0.5132 | Yes | ||
| 92 | CXXC6 | 16185 | -2.984 | -0.5066 | Yes | ||
| 93 | ZFYVE26 | 16239 | -3.077 | -0.5022 | Yes | ||
| 94 | CUGBP2 | 16257 | -3.116 | -0.4958 | Yes | ||
| 95 | GNG5 | 16266 | -3.132 | -0.4888 | Yes | ||
| 96 | ENAH | 16424 | -3.410 | -0.4893 | Yes | ||
| 97 | TPM3 | 16450 | -3.455 | -0.4825 | Yes | ||
| 98 | CTDSP1 | 16610 | -3.781 | -0.4821 | Yes | ||
| 99 | ARHGEF18 | 16663 | -3.885 | -0.4758 | Yes | ||
| 100 | RAB8B | 16801 | -4.206 | -0.4733 | Yes | ||
| 101 | RPS6KA3 | 16838 | -4.270 | -0.4651 | Yes | ||
| 102 | USP47 | 16859 | -4.308 | -0.4560 | Yes | ||
| 103 | EIF2C2 | 16911 | -4.445 | -0.4483 | Yes | ||
| 104 | MXD4 | 16968 | -4.544 | -0.4406 | Yes | ||
| 105 | PPP2R5C | 17001 | -4.612 | -0.4314 | Yes | ||
| 106 | PFN2 | 17060 | -4.747 | -0.4234 | Yes | ||
| 107 | ZNF289 | 17077 | -4.803 | -0.4129 | Yes | ||
| 108 | BNIP3L | 17145 | -5.009 | -0.4047 | Yes | ||
| 109 | ING3 | 17233 | -5.277 | -0.3969 | Yes | ||
| 110 | DGCR2 | 17274 | -5.408 | -0.3863 | Yes | ||
| 111 | NR3C1 | 17288 | -5.464 | -0.3741 | Yes | ||
| 112 | PIM1 | 17322 | -5.526 | -0.3629 | Yes | ||
| 113 | FOXP1 | 17514 | -6.087 | -0.3588 | Yes | ||
| 114 | MBNL1 | 17740 | -6.866 | -0.3548 | Yes | ||
| 115 | RHOBTB1 | 17860 | -7.461 | -0.3436 | Yes | ||
| 116 | TRAM1 | 17881 | -7.565 | -0.3268 | Yes | ||
| 117 | PSEN2 | 17891 | -7.601 | -0.3094 | Yes | ||
| 118 | TTC7B | 18079 | -8.789 | -0.2988 | Yes | ||
| 119 | PRKCA | 18108 | -9.028 | -0.2790 | Yes | ||
| 120 | TCF12 | 18161 | -9.536 | -0.2593 | Yes | ||
| 121 | FOXO1A | 18314 | -11.014 | -0.2415 | Yes | ||
| 122 | CSNK1G3 | 18342 | -11.482 | -0.2158 | Yes | ||
| 123 | BTG1 | 18363 | -11.850 | -0.1889 | Yes | ||
| 124 | DAP | 18394 | -12.647 | -0.1607 | Yes | ||
| 125 | MEF2C | 18457 | -14.583 | -0.1296 | Yes | ||
| 126 | TLE4 | 18471 | -15.081 | -0.0947 | Yes | ||
| 127 | DUSP10 | 18536 | -20.147 | -0.0506 | Yes | ||
| 128 | EGR1 | 18550 | -23.249 | 0.0036 | Yes |