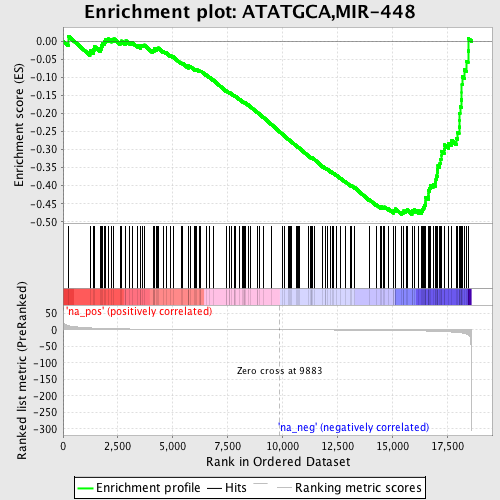
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATATGCA,MIR-448 |
| Enrichment Score (ES) | -0.47973877 |
| Normalized Enrichment Score (NES) | -1.5591258 |
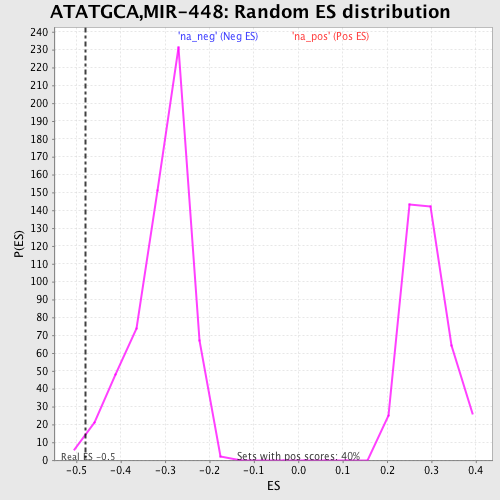
| Nominal p-value | 0.01 |
| FDR q-value | 0.977134 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STK4 | 239 | 12.046 | 0.0144 | No | ||
| 2 | LRRIQ2 | 1226 | 5.627 | -0.0263 | No | ||
| 3 | PRKAR2B | 1389 | 5.093 | -0.0235 | No | ||
| 4 | DTNA | 1413 | 5.021 | -0.0134 | No | ||
| 5 | TPP2 | 1692 | 4.319 | -0.0186 | No | ||
| 6 | FUSIP1 | 1750 | 4.189 | -0.0122 | No | ||
| 7 | RNGTT | 1807 | 4.044 | -0.0061 | No | ||
| 8 | PTBP1 | 1876 | 3.891 | -0.0009 | No | ||
| 9 | PCDHGB1 | 1941 | 3.773 | 0.0042 | No | ||
| 10 | BCL2 | 2046 | 3.584 | 0.0067 | No | ||
| 11 | RANBP2 | 2207 | 3.299 | 0.0055 | No | ||
| 12 | PCDHGC5 | 2318 | 3.131 | 0.0066 | No | ||
| 13 | HNRPA2B1 | 2596 | 2.786 | -0.0021 | No | ||
| 14 | E2F3 | 2656 | 2.722 | 0.0009 | No | ||
| 15 | UNC5A | 2832 | 2.537 | -0.0028 | No | ||
| 16 | AZIN1 | 2865 | 2.507 | 0.0012 | No | ||
| 17 | PCDHGA3 | 3030 | 2.365 | -0.0024 | No | ||
| 18 | GLULD1 | 3155 | 2.248 | -0.0040 | No | ||
| 19 | WDFY3 | 3381 | 2.086 | -0.0114 | No | ||
| 20 | KPNA3 | 3532 | 1.989 | -0.0151 | No | ||
| 21 | SCN5A | 3535 | 1.987 | -0.0107 | No | ||
| 22 | KLHL5 | 3620 | 1.931 | -0.0108 | No | ||
| 23 | AMMECR1 | 3688 | 1.891 | -0.0102 | No | ||
| 24 | PPM2C | 4096 | 1.692 | -0.0284 | No | ||
| 25 | FMR1 | 4115 | 1.684 | -0.0255 | No | ||
| 26 | VLDLR | 4148 | 1.667 | -0.0235 | No | ||
| 27 | PCDH8 | 4164 | 1.659 | -0.0205 | No | ||
| 28 | ATP11A | 4252 | 1.610 | -0.0216 | No | ||
| 29 | NAV1 | 4282 | 1.597 | -0.0195 | No | ||
| 30 | KCNH7 | 4328 | 1.580 | -0.0184 | No | ||
| 31 | IGF1R | 4582 | 1.477 | -0.0288 | No | ||
| 32 | SH3GL2 | 4693 | 1.432 | -0.0315 | No | ||
| 33 | KCTD15 | 4913 | 1.350 | -0.0403 | No | ||
| 34 | GNAI1 | 5017 | 1.320 | -0.0429 | No | ||
| 35 | DNM3 | 5385 | 1.191 | -0.0600 | No | ||
| 36 | GLCE | 5442 | 1.176 | -0.0604 | No | ||
| 37 | KCNB1 | 5699 | 1.093 | -0.0718 | No | ||
| 38 | COLEC12 | 5700 | 1.093 | -0.0693 | No | ||
| 39 | SYN2 | 5707 | 1.092 | -0.0672 | No | ||
| 40 | KCNMB2 | 5810 | 1.060 | -0.0703 | No | ||
| 41 | PCDHGA9 | 6005 | 1.007 | -0.0785 | No | ||
| 42 | NLGN1 | 6024 | 0.997 | -0.0772 | No | ||
| 43 | NTF3 | 6066 | 0.983 | -0.0772 | No | ||
| 44 | SMURF1 | 6195 | 0.944 | -0.0820 | No | ||
| 45 | DBC1 | 6249 | 0.927 | -0.0828 | No | ||
| 46 | GRIK2 | 6281 | 0.917 | -0.0824 | No | ||
| 47 | AFF2 | 6515 | 0.849 | -0.0931 | No | ||
| 48 | CRY1 | 6670 | 0.807 | -0.0996 | No | ||
| 49 | FBXW11 | 6840 | 0.764 | -0.1070 | No | ||
| 50 | DOCK9 | 7427 | 0.609 | -0.1374 | No | ||
| 51 | RAB2B | 7468 | 0.598 | -0.1382 | No | ||
| 52 | PCDHGA2 | 7595 | 0.565 | -0.1437 | No | ||
| 53 | BRP44L | 7599 | 0.565 | -0.1426 | No | ||
| 54 | PCDHGA8 | 7661 | 0.549 | -0.1447 | No | ||
| 55 | APPBP2 | 7811 | 0.510 | -0.1516 | No | ||
| 56 | AHR | 7816 | 0.507 | -0.1507 | No | ||
| 57 | HECTD2 | 7865 | 0.492 | -0.1521 | No | ||
| 58 | DNAJB11 | 8035 | 0.452 | -0.1603 | No | ||
| 59 | PCDHGA5 | 8188 | 0.412 | -0.1676 | No | ||
| 60 | PCDHGA6 | 8229 | 0.405 | -0.1688 | No | ||
| 61 | PRKAB2 | 8272 | 0.393 | -0.1702 | No | ||
| 62 | GDF6 | 8273 | 0.393 | -0.1693 | No | ||
| 63 | DOC2A | 8318 | 0.382 | -0.1708 | No | ||
| 64 | PCDHGC4 | 8428 | 0.354 | -0.1759 | No | ||
| 65 | PCDHGB7 | 8560 | 0.322 | -0.1823 | No | ||
| 66 | ATXN3 | 8846 | 0.253 | -0.1972 | No | ||
| 67 | ZCCHC2 | 8872 | 0.248 | -0.1980 | No | ||
| 68 | PCDHGB5 | 8932 | 0.232 | -0.2006 | No | ||
| 69 | SOX11 | 9133 | 0.186 | -0.2110 | No | ||
| 70 | PCDHGA7 | 9488 | 0.095 | -0.2300 | No | ||
| 71 | ZFHX4 | 9995 | -0.031 | -0.2574 | No | ||
| 72 | TCEAL1 | 10101 | -0.057 | -0.2629 | No | ||
| 73 | ANK2 | 10261 | -0.094 | -0.2713 | No | ||
| 74 | PURA | 10326 | -0.109 | -0.2745 | No | ||
| 75 | RAB11FIP2 | 10350 | -0.116 | -0.2755 | No | ||
| 76 | SP3 | 10431 | -0.137 | -0.2795 | No | ||
| 77 | EPHA4 | 10624 | -0.186 | -0.2895 | No | ||
| 78 | PCDHGB6 | 10692 | -0.206 | -0.2927 | No | ||
| 79 | TMSB4X | 10728 | -0.217 | -0.2941 | No | ||
| 80 | CLCN5 | 10771 | -0.226 | -0.2959 | No | ||
| 81 | DAPK1 | 11174 | -0.339 | -0.3169 | No | ||
| 82 | TBX3 | 11295 | -0.372 | -0.3225 | No | ||
| 83 | ADAM10 | 11302 | -0.374 | -0.3220 | No | ||
| 84 | XYLT2 | 11337 | -0.380 | -0.3230 | No | ||
| 85 | GABRB3 | 11349 | -0.384 | -0.3227 | No | ||
| 86 | PCDH9 | 11444 | -0.408 | -0.3269 | No | ||
| 87 | RORB | 11844 | -0.516 | -0.3473 | No | ||
| 88 | TOPORS | 11953 | -0.547 | -0.3519 | No | ||
| 89 | HOXA11 | 11964 | -0.549 | -0.3512 | No | ||
| 90 | PCDHGA4 | 12049 | -0.572 | -0.3545 | No | ||
| 91 | KLF5 | 12208 | -0.624 | -0.3616 | No | ||
| 92 | SF3B1 | 12288 | -0.645 | -0.3644 | No | ||
| 93 | GPHN | 12336 | -0.660 | -0.3655 | No | ||
| 94 | PCDHGA1 | 12468 | -0.702 | -0.3710 | No | ||
| 95 | SLC7A11 | 12658 | -0.768 | -0.3795 | No | ||
| 96 | CENTG2 | 12863 | -0.829 | -0.3887 | No | ||
| 97 | SS18 | 13093 | -0.908 | -0.3990 | No | ||
| 98 | PCDHGA10 | 13155 | -0.933 | -0.4002 | No | ||
| 99 | CPEB2 | 13280 | -0.975 | -0.4047 | No | ||
| 100 | EPHB2 | 13966 | -1.251 | -0.4390 | No | ||
| 101 | DACT1 | 14277 | -1.409 | -0.4526 | No | ||
| 102 | CASP4 | 14468 | -1.514 | -0.4594 | No | ||
| 103 | THRAP2 | 14513 | -1.536 | -0.4583 | No | ||
| 104 | OTX2 | 14612 | -1.587 | -0.4601 | No | ||
| 105 | PAPPA | 14660 | -1.617 | -0.4589 | No | ||
| 106 | PLEKHA6 | 14823 | -1.700 | -0.4638 | No | ||
| 107 | NRIP1 | 15047 | -1.853 | -0.4717 | No | ||
| 108 | PCDHGB2 | 15074 | -1.867 | -0.4689 | No | ||
| 109 | MYH3 | 15140 | -1.906 | -0.4681 | No | ||
| 110 | CNTN4 | 15151 | -1.913 | -0.4643 | No | ||
| 111 | WRNIP1 | 15437 | -2.147 | -0.4749 | Yes | ||
| 112 | BICD2 | 15497 | -2.200 | -0.4731 | Yes | ||
| 113 | BNC2 | 15533 | -2.244 | -0.4699 | Yes | ||
| 114 | APBB2 | 15628 | -2.334 | -0.4697 | Yes | ||
| 115 | BACH2 | 15673 | -2.376 | -0.4667 | Yes | ||
| 116 | ELL2 | 15903 | -2.618 | -0.4731 | Yes | ||
| 117 | CXCL12 | 15918 | -2.642 | -0.4679 | Yes | ||
| 118 | RNF19 | 16014 | -2.756 | -0.4668 | Yes | ||
| 119 | YTHDF3 | 16189 | -2.994 | -0.4694 | Yes | ||
| 120 | SIRT1 | 16332 | -3.238 | -0.4698 | Yes | ||
| 121 | GPM6A | 16380 | -3.304 | -0.4648 | Yes | ||
| 122 | PCDHGB4 | 16426 | -3.413 | -0.4595 | Yes | ||
| 123 | ARMC8 | 16471 | -3.494 | -0.4540 | Yes | ||
| 124 | CLK1 | 16509 | -3.576 | -0.4479 | Yes | ||
| 125 | ELF2 | 16514 | -3.590 | -0.4399 | Yes | ||
| 126 | ADD1 | 16516 | -3.595 | -0.4318 | Yes | ||
| 127 | LYST | 16648 | -3.851 | -0.4302 | Yes | ||
| 128 | FBXL17 | 16656 | -3.866 | -0.4218 | Yes | ||
| 129 | KCTD9 | 16659 | -3.877 | -0.4131 | Yes | ||
| 130 | FBXO33 | 16705 | -3.970 | -0.4066 | Yes | ||
| 131 | SERTAD2 | 16751 | -4.090 | -0.3997 | Yes | ||
| 132 | ARID2 | 16861 | -4.310 | -0.3958 | Yes | ||
| 133 | BRD1 | 16962 | -4.533 | -0.3910 | Yes | ||
| 134 | MAP2K6 | 16991 | -4.589 | -0.3821 | Yes | ||
| 135 | DLGAP4 | 17018 | -4.646 | -0.3730 | Yes | ||
| 136 | NCOA2 | 17052 | -4.724 | -0.3640 | Yes | ||
| 137 | PFN2 | 17060 | -4.747 | -0.3536 | Yes | ||
| 138 | RAP2C | 17082 | -4.821 | -0.3438 | Yes | ||
| 139 | CACNA1C | 17173 | -5.100 | -0.3371 | Yes | ||
| 140 | DYNLT3 | 17186 | -5.126 | -0.3262 | Yes | ||
| 141 | RAB21 | 17229 | -5.266 | -0.3165 | Yes | ||
| 142 | ING3 | 17233 | -5.277 | -0.3047 | Yes | ||
| 143 | WTAP | 17367 | -5.699 | -0.2990 | Yes | ||
| 144 | DPP4 | 17385 | -5.739 | -0.2869 | Yes | ||
| 145 | FBXO28 | 17567 | -6.295 | -0.2824 | Yes | ||
| 146 | BCL11A | 17700 | -6.753 | -0.2742 | Yes | ||
| 147 | AZI2 | 17919 | -7.745 | -0.2685 | Yes | ||
| 148 | ARNTL | 17981 | -8.187 | -0.2532 | Yes | ||
| 149 | SH3KBP1 | 18046 | -8.546 | -0.2373 | Yes | ||
| 150 | ETV1 | 18059 | -8.622 | -0.2184 | Yes | ||
| 151 | MBNL2 | 18065 | -8.653 | -0.1990 | Yes | ||
| 152 | SOCS5 | 18091 | -8.885 | -0.1802 | Yes | ||
| 153 | KLF13 | 18147 | -9.369 | -0.1619 | Yes | ||
| 154 | PELI1 | 18177 | -9.668 | -0.1416 | Yes | ||
| 155 | WDR22 | 18179 | -9.670 | -0.1197 | Yes | ||
| 156 | BTG3 | 18191 | -9.860 | -0.0979 | Yes | ||
| 157 | UBL3 | 18284 | -10.612 | -0.0788 | Yes | ||
| 158 | KRAS | 18389 | -12.525 | -0.0560 | Yes | ||
| 159 | MEF2C | 18457 | -14.583 | -0.0266 | Yes | ||
| 160 | CAP1 | 18484 | -15.486 | 0.0072 | Yes |