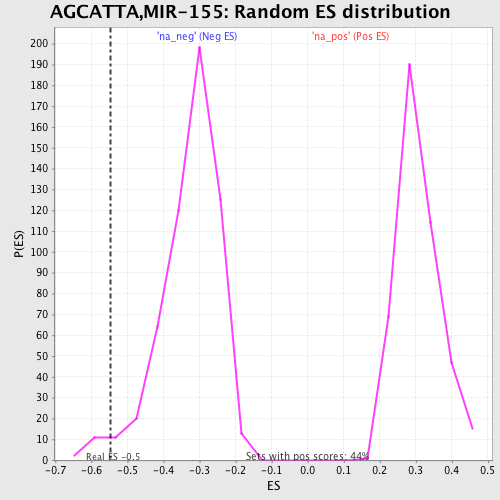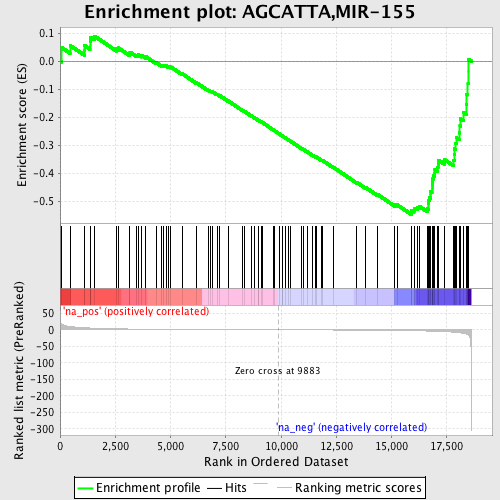
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | AGCATTA,MIR-155 |
| Enrichment Score (ES) | -0.5471398 |
| Normalized Enrichment Score (NES) | -1.6675057 |
| Nominal p-value | 0.023049645 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TIMM8A | 60 | 18.225 | 0.0492 | No | ||
| 2 | BCAP29 | 451 | 9.810 | 0.0563 | No | ||
| 3 | PICALM | 1096 | 6.067 | 0.0390 | No | ||
| 4 | KCNN3 | 1102 | 6.045 | 0.0561 | No | ||
| 5 | YWHAZ | 1354 | 5.245 | 0.0577 | No | ||
| 6 | TRIM2 | 1355 | 5.238 | 0.0727 | No | ||
| 7 | BCAT1 | 1384 | 5.100 | 0.0859 | No | ||
| 8 | PSIP1 | 1561 | 4.602 | 0.0896 | No | ||
| 9 | YWHAE | 2553 | 2.833 | 0.0443 | No | ||
| 10 | CHD7 | 2640 | 2.742 | 0.0475 | No | ||
| 11 | GPM6B | 3148 | 2.255 | 0.0266 | No | ||
| 12 | WEE1 | 3159 | 2.243 | 0.0325 | No | ||
| 13 | MAP3K10 | 3461 | 2.034 | 0.0221 | No | ||
| 14 | SKI | 3538 | 1.986 | 0.0237 | No | ||
| 15 | QKI | 3701 | 1.887 | 0.0204 | No | ||
| 16 | CEBPB | 3882 | 1.792 | 0.0159 | No | ||
| 17 | PKIA | 4358 | 1.563 | -0.0053 | No | ||
| 18 | ITK | 4603 | 1.466 | -0.0143 | No | ||
| 19 | SLC39A10 | 4686 | 1.435 | -0.0145 | No | ||
| 20 | GPR85 | 4793 | 1.398 | -0.0163 | No | ||
| 21 | MYO1D | 4893 | 1.358 | -0.0177 | No | ||
| 22 | CSF1R | 5016 | 1.320 | -0.0205 | No | ||
| 23 | HIF1A | 5533 | 1.149 | -0.0450 | No | ||
| 24 | LRP1B | 6164 | 0.955 | -0.0763 | No | ||
| 25 | CDC73 | 6721 | 0.793 | -0.1041 | No | ||
| 26 | PTPN2 | 6823 | 0.768 | -0.1073 | No | ||
| 27 | SOX1 | 6880 | 0.754 | -0.1082 | No | ||
| 28 | SGCZ | 7101 | 0.698 | -0.1180 | No | ||
| 29 | SEMA5A | 7228 | 0.664 | -0.1229 | No | ||
| 30 | BOC | 7605 | 0.563 | -0.1416 | No | ||
| 31 | GDF6 | 8273 | 0.393 | -0.1765 | No | ||
| 32 | HIVEP2 | 8358 | 0.371 | -0.1800 | No | ||
| 33 | NFIX | 8641 | 0.303 | -0.1943 | No | ||
| 34 | RCN2 | 8819 | 0.259 | -0.2031 | No | ||
| 35 | ACTA1 | 8984 | 0.219 | -0.2114 | No | ||
| 36 | ACVR2A | 8986 | 0.218 | -0.2108 | No | ||
| 37 | SOX11 | 9133 | 0.186 | -0.2181 | No | ||
| 38 | SYPL1 | 9165 | 0.180 | -0.2193 | No | ||
| 39 | BCORL1 | 9667 | 0.057 | -0.2462 | No | ||
| 40 | CAB39 | 9688 | 0.052 | -0.2471 | No | ||
| 41 | SERAC1 | 9916 | -0.009 | -0.2593 | No | ||
| 42 | ZIC3 | 10075 | -0.051 | -0.2677 | No | ||
| 43 | IKBKE | 10076 | -0.051 | -0.2676 | No | ||
| 44 | OGN | 10205 | -0.079 | -0.2743 | No | ||
| 45 | RAB11FIP2 | 10350 | -0.116 | -0.2817 | No | ||
| 46 | SP3 | 10431 | -0.137 | -0.2856 | No | ||
| 47 | CTLA4 | 10947 | -0.274 | -0.3126 | No | ||
| 48 | NDFIP1 | 11024 | -0.295 | -0.3159 | No | ||
| 49 | TOMM20 | 11181 | -0.341 | -0.3233 | No | ||
| 50 | PCDH9 | 11444 | -0.408 | -0.3363 | No | ||
| 51 | DYNC1I1 | 11537 | -0.437 | -0.3400 | No | ||
| 52 | DNAJB7 | 11592 | -0.453 | -0.3416 | No | ||
| 53 | UBE2D3 | 11842 | -0.516 | -0.3536 | No | ||
| 54 | SCG2 | 11883 | -0.528 | -0.3542 | No | ||
| 55 | EDG1 | 12362 | -0.670 | -0.3781 | No | ||
| 56 | MARCH7 | 13422 | -1.026 | -0.4324 | No | ||
| 57 | FGF7 | 13825 | -1.190 | -0.4506 | No | ||
| 58 | MYO10 | 14345 | -1.444 | -0.4745 | No | ||
| 59 | CNTN4 | 15151 | -1.913 | -0.5125 | No | ||
| 60 | UPP2 | 15250 | -1.975 | -0.5121 | No | ||
| 61 | SMARCA4 | 15900 | -2.615 | -0.5396 | Yes | ||
| 62 | ELL2 | 15903 | -2.618 | -0.5322 | Yes | ||
| 63 | NFAT5 | 16018 | -2.763 | -0.5304 | Yes | ||
| 64 | SGK3 | 16055 | -2.808 | -0.5243 | Yes | ||
| 65 | ARL5B | 16182 | -2.980 | -0.5225 | Yes | ||
| 66 | CUGBP2 | 16257 | -3.116 | -0.5175 | Yes | ||
| 67 | MYB | 16630 | -3.818 | -0.5266 | Yes | ||
| 68 | STRN3 | 16678 | -3.910 | -0.5179 | Yes | ||
| 69 | BACH1 | 16679 | -3.910 | -0.5067 | Yes | ||
| 70 | CSNK1G2 | 16688 | -3.925 | -0.4958 | Yes | ||
| 71 | FBXO33 | 16705 | -3.970 | -0.4853 | Yes | ||
| 72 | MAP3K14 | 16772 | -4.141 | -0.4769 | Yes | ||
| 73 | RAB1A | 16774 | -4.145 | -0.4650 | Yes | ||
| 74 | SDCBP | 16834 | -4.265 | -0.4560 | Yes | ||
| 75 | RPS6KA3 | 16838 | -4.270 | -0.4438 | Yes | ||
| 76 | INPP5D | 16846 | -4.291 | -0.4319 | Yes | ||
| 77 | ARID2 | 16861 | -4.310 | -0.4202 | Yes | ||
| 78 | ETS1 | 16882 | -4.362 | -0.4088 | Yes | ||
| 79 | MEIS1 | 16952 | -4.525 | -0.3995 | Yes | ||
| 80 | BRD1 | 16962 | -4.533 | -0.3869 | Yes | ||
| 81 | MECP2 | 17092 | -4.861 | -0.3799 | Yes | ||
| 82 | RAB6A | 17108 | -4.921 | -0.3666 | Yes | ||
| 83 | H3F3A | 17131 | -4.977 | -0.3534 | Yes | ||
| 84 | DNAJB1 | 17389 | -5.745 | -0.3508 | Yes | ||
| 85 | SPI1 | 17810 | -7.163 | -0.3529 | Yes | ||
| 86 | CD47 | 17841 | -7.316 | -0.3334 | Yes | ||
| 87 | KBTBD2 | 17854 | -7.427 | -0.3127 | Yes | ||
| 88 | SSH2 | 17889 | -7.595 | -0.2927 | Yes | ||
| 89 | CUTL1 | 17932 | -7.841 | -0.2724 | Yes | ||
| 90 | MAP3K7IP2 | 18062 | -8.635 | -0.2545 | Yes | ||
| 91 | CARHSP1 | 18097 | -8.935 | -0.2307 | Yes | ||
| 92 | SOCS1 | 18119 | -9.137 | -0.2055 | Yes | ||
| 93 | SATB1 | 18248 | -10.370 | -0.1826 | Yes | ||
| 94 | KRAS | 18389 | -12.525 | -0.1541 | Yes | ||
| 95 | MIDN | 18407 | -12.955 | -0.1178 | Yes | ||
| 96 | SLA | 18460 | -14.650 | -0.0784 | Yes | ||
| 97 | TLE4 | 18471 | -15.081 | -0.0356 | Yes | ||
| 98 | HBP1 | 18472 | -15.082 | 0.0078 | Yes |