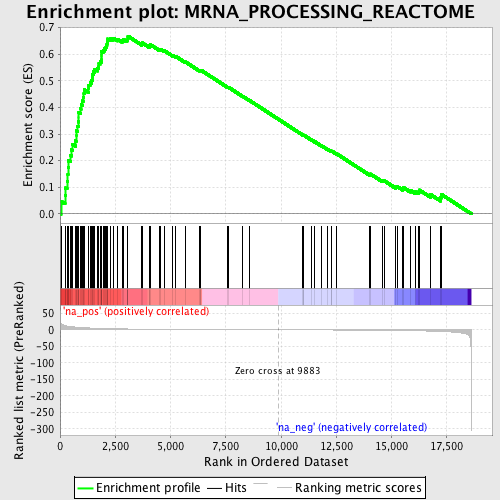
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MRNA_PROCESSING_REACTOME |
| Enrichment Score (ES) | 0.66755915 |
| Normalized Enrichment Score (NES) | 2.1968412 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SRPK1 | 41 | 19.475 | 0.0472 | Yes | ||
| 2 | SNRPD1 | 224 | 12.313 | 0.0687 | Yes | ||
| 3 | U2AF1 | 252 | 11.890 | 0.0974 | Yes | ||
| 4 | SNRPF | 315 | 11.120 | 0.1223 | Yes | ||
| 5 | SFRS6 | 352 | 10.664 | 0.1474 | Yes | ||
| 6 | LSM7 | 361 | 10.583 | 0.1739 | Yes | ||
| 7 | SNRPD3 | 380 | 10.453 | 0.1995 | Yes | ||
| 8 | PHF5A | 464 | 9.691 | 0.2196 | Yes | ||
| 9 | SNRPA1 | 509 | 9.418 | 0.2411 | Yes | ||
| 10 | CPSF1 | 551 | 9.092 | 0.2620 | Yes | ||
| 11 | XRN2 | 700 | 8.032 | 0.2744 | Yes | ||
| 12 | CSTF3 | 726 | 7.879 | 0.2931 | Yes | ||
| 13 | TXNL4A | 731 | 7.838 | 0.3127 | Yes | ||
| 14 | PRPF3 | 779 | 7.584 | 0.3295 | Yes | ||
| 15 | SNRPG | 825 | 7.378 | 0.3458 | Yes | ||
| 16 | SF3A3 | 843 | 7.271 | 0.3633 | Yes | ||
| 17 | SNRPD2 | 844 | 7.271 | 0.3818 | Yes | ||
| 18 | NCBP2 | 918 | 6.876 | 0.3953 | Yes | ||
| 19 | DHX9 | 948 | 6.750 | 0.4109 | Yes | ||
| 20 | PRPF4 | 1003 | 6.455 | 0.4244 | Yes | ||
| 21 | RNPS1 | 1043 | 6.318 | 0.4383 | Yes | ||
| 22 | CPSF4 | 1078 | 6.146 | 0.4521 | Yes | ||
| 23 | CSTF1 | 1106 | 6.017 | 0.4659 | Yes | ||
| 24 | CUGBP1 | 1281 | 5.467 | 0.4704 | Yes | ||
| 25 | DNAJC8 | 1290 | 5.444 | 0.4838 | Yes | ||
| 26 | DDX1 | 1356 | 5.229 | 0.4935 | Yes | ||
| 27 | SFRS2 | 1426 | 4.971 | 0.5024 | Yes | ||
| 28 | SNRPA | 1459 | 4.879 | 0.5131 | Yes | ||
| 29 | SFRS5 | 1470 | 4.834 | 0.5248 | Yes | ||
| 30 | SNRPB | 1510 | 4.709 | 0.5347 | Yes | ||
| 31 | RNMT | 1565 | 4.588 | 0.5434 | Yes | ||
| 32 | DDX20 | 1695 | 4.314 | 0.5474 | Yes | ||
| 33 | FUSIP1 | 1750 | 4.189 | 0.5551 | Yes | ||
| 34 | LSM2 | 1757 | 4.167 | 0.5654 | Yes | ||
| 35 | RNGTT | 1807 | 4.044 | 0.5730 | Yes | ||
| 36 | POLR2A | 1865 | 3.916 | 0.5799 | Yes | ||
| 37 | SNRP70 | 1871 | 3.900 | 0.5895 | Yes | ||
| 38 | HNRPAB | 1873 | 3.893 | 0.5993 | Yes | ||
| 39 | PTBP1 | 1876 | 3.891 | 0.6091 | Yes | ||
| 40 | PRPF8 | 1947 | 3.762 | 0.6149 | Yes | ||
| 41 | FUS | 2012 | 3.648 | 0.6207 | Yes | ||
| 42 | SF3A1 | 2069 | 3.541 | 0.6267 | Yes | ||
| 43 | HNRPH1 | 2090 | 3.508 | 0.6345 | Yes | ||
| 44 | HNRPA3 | 2123 | 3.451 | 0.6415 | Yes | ||
| 45 | CSTF2 | 2156 | 3.396 | 0.6484 | Yes | ||
| 46 | RBM17 | 2161 | 3.389 | 0.6568 | Yes | ||
| 47 | HNRPU | 2293 | 3.169 | 0.6578 | Yes | ||
| 48 | SF3B5 | 2393 | 3.025 | 0.6601 | Yes | ||
| 49 | HNRPA2B1 | 2596 | 2.786 | 0.6563 | Yes | ||
| 50 | PABPN1 | 2805 | 2.559 | 0.6515 | Yes | ||
| 51 | DHX15 | 2860 | 2.511 | 0.6550 | Yes | ||
| 52 | PPM1G | 3028 | 2.366 | 0.6520 | Yes | ||
| 53 | HNRPC | 3064 | 2.321 | 0.6560 | Yes | ||
| 54 | CPSF3 | 3069 | 2.318 | 0.6617 | Yes | ||
| 55 | SFRS9 | 3070 | 2.317 | 0.6676 | Yes | ||
| 56 | U2AF2 | 3699 | 1.888 | 0.6384 | No | ||
| 57 | CLK4 | 3719 | 1.873 | 0.6422 | No | ||
| 58 | SFRS10 | 4036 | 1.720 | 0.6295 | No | ||
| 59 | SF3B2 | 4038 | 1.720 | 0.6338 | No | ||
| 60 | HNRPL | 4077 | 1.699 | 0.6361 | No | ||
| 61 | DICER1 | 4503 | 1.508 | 0.6169 | No | ||
| 62 | DHX38 | 4546 | 1.492 | 0.6185 | No | ||
| 63 | NUDT21 | 4724 | 1.421 | 0.6125 | No | ||
| 64 | SFRS16 | 5102 | 1.288 | 0.5954 | No | ||
| 65 | SF3A2 | 5229 | 1.247 | 0.5918 | No | ||
| 66 | DHX16 | 5670 | 1.102 | 0.5708 | No | ||
| 67 | PTBP2 | 6313 | 0.909 | 0.5385 | No | ||
| 68 | SFRS7 | 6339 | 0.900 | 0.5394 | No | ||
| 69 | SNRPE | 6369 | 0.891 | 0.5401 | No | ||
| 70 | NONO | 7582 | 0.569 | 0.4761 | No | ||
| 71 | CSTF2T | 7619 | 0.560 | 0.4756 | No | ||
| 72 | HNRPD | 8252 | 0.397 | 0.4424 | No | ||
| 73 | DHX8 | 8559 | 0.322 | 0.4267 | No | ||
| 74 | SF4 | 10990 | -0.284 | 0.2962 | No | ||
| 75 | PRPF4B | 11006 | -0.290 | 0.2962 | No | ||
| 76 | COL2A1 | 11018 | -0.293 | 0.2963 | No | ||
| 77 | HNRPH2 | 11393 | -0.398 | 0.2771 | No | ||
| 78 | SF3B4 | 11535 | -0.436 | 0.2706 | No | ||
| 79 | METTL3 | 11831 | -0.513 | 0.2560 | No | ||
| 80 | HNRPR | 12118 | -0.592 | 0.2421 | No | ||
| 81 | SUPT5H | 12261 | -0.638 | 0.2360 | No | ||
| 82 | SF3B1 | 12288 | -0.645 | 0.2362 | No | ||
| 83 | SFRS14 | 12513 | -0.715 | 0.2260 | No | ||
| 84 | SFRS8 | 14016 | -1.277 | 0.1481 | No | ||
| 85 | SNRPB2 | 14044 | -1.295 | 0.1499 | No | ||
| 86 | SRPK2 | 14580 | -1.572 | 0.1250 | No | ||
| 87 | NXF1 | 14677 | -1.625 | 0.1240 | No | ||
| 88 | SNRPN | 15192 | -1.940 | 0.1012 | No | ||
| 89 | PAPOLA | 15251 | -1.976 | 0.1030 | No | ||
| 90 | GIPC1 | 15512 | -2.216 | 0.0946 | No | ||
| 91 | SPOP | 15523 | -2.233 | 0.0998 | No | ||
| 92 | CLK2 | 15852 | -2.558 | 0.0885 | No | ||
| 93 | SRRM1 | 16093 | -2.852 | 0.0828 | No | ||
| 94 | CPSF2 | 16237 | -3.076 | 0.0829 | No | ||
| 95 | CUGBP2 | 16257 | -3.116 | 0.0898 | No | ||
| 96 | DDIT3 | 16760 | -4.120 | 0.0732 | No | ||
| 97 | CD2BP2 | 17223 | -5.255 | 0.0616 | No | ||
| 98 | RBM5 | 17260 | -5.361 | 0.0732 | No |