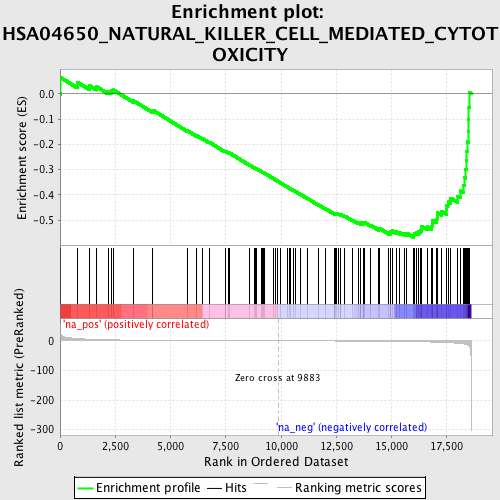
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04650_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY |
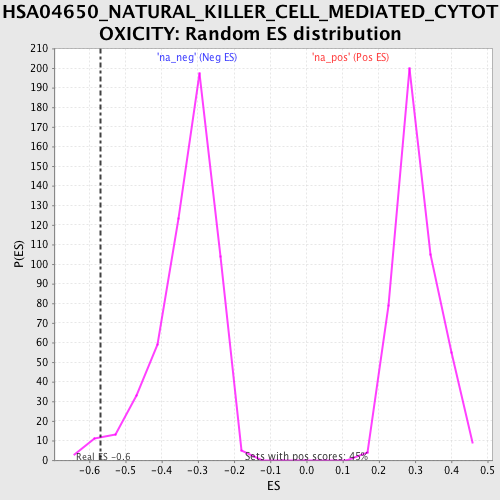
| Enrichment Score (ES) | -0.567934 |
| Normalized Enrichment Score (NES) | -1.7083532 |
| Nominal p-value | 0.016423358 |
| FDR q-value | 0.34066048 |
| FWER p-Value | 0.995 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BID | 24 | 21.976 | 0.0641 | No | ||
| 2 | PIK3CB | 780 | 7.582 | 0.0459 | No | ||
| 3 | CD48 | 1320 | 5.342 | 0.0327 | No | ||
| 4 | PIK3R3 | 1649 | 4.388 | 0.0281 | No | ||
| 5 | HCST | 2180 | 3.358 | 0.0095 | No | ||
| 6 | CASP3 | 2316 | 3.134 | 0.0115 | No | ||
| 7 | PTPN11 | 2411 | 3.002 | 0.0154 | No | ||
| 8 | IFNA1 | 3323 | 2.127 | -0.0275 | No | ||
| 9 | SHC1 | 4178 | 1.653 | -0.0687 | No | ||
| 10 | IFNA7 | 4188 | 1.647 | -0.0643 | No | ||
| 11 | IFNA2 | 5761 | 1.074 | -0.1459 | No | ||
| 12 | HRAS | 6193 | 0.946 | -0.1664 | No | ||
| 13 | TNFRSF10B | 6430 | 0.873 | -0.1765 | No | ||
| 14 | FCER1G | 6747 | 0.788 | -0.1913 | No | ||
| 15 | KIR3DL1 | 7465 | 0.599 | -0.2282 | No | ||
| 16 | KLRK1 | 7490 | 0.591 | -0.2277 | No | ||
| 17 | PIK3R2 | 7604 | 0.563 | -0.2321 | No | ||
| 18 | IFNB1 | 7668 | 0.547 | -0.2339 | No | ||
| 19 | IFNA6 | 8566 | 0.320 | -0.2814 | No | ||
| 20 | TNFSF10 | 8784 | 0.267 | -0.2923 | No | ||
| 21 | IFNA14 | 8836 | 0.256 | -0.2943 | No | ||
| 22 | PIK3R5 | 8866 | 0.250 | -0.2951 | No | ||
| 23 | CD247 | 9117 | 0.190 | -0.3081 | No | ||
| 24 | PAK1 | 9149 | 0.183 | -0.3092 | No | ||
| 25 | NRAS | 9203 | 0.170 | -0.3116 | No | ||
| 26 | SHC3 | 9261 | 0.155 | -0.3142 | No | ||
| 27 | KLRC1 | 9652 | 0.061 | -0.3350 | No | ||
| 28 | PRF1 | 9750 | 0.036 | -0.3402 | No | ||
| 29 | PPP3R2 | 9840 | 0.013 | -0.3449 | No | ||
| 30 | SH2D1A | 9972 | -0.024 | -0.3519 | No | ||
| 31 | RAC3 | 10299 | -0.101 | -0.3692 | No | ||
| 32 | LAT | 10401 | -0.130 | -0.3743 | No | ||
| 33 | NCR1 | 10448 | -0.142 | -0.3764 | No | ||
| 34 | FCGR3A | 10560 | -0.170 | -0.3819 | No | ||
| 35 | SH2D1B | 10640 | -0.192 | -0.3856 | No | ||
| 36 | NFATC4 | 10877 | -0.257 | -0.3975 | No | ||
| 37 | NFATC2 | 11197 | -0.344 | -0.4137 | No | ||
| 38 | LCP2 | 11709 | -0.483 | -0.4399 | No | ||
| 39 | IFNA13 | 12022 | -0.564 | -0.4550 | No | ||
| 40 | TYROBP | 12407 | -0.682 | -0.4737 | No | ||
| 41 | MAP2K2 | 12431 | -0.690 | -0.4729 | No | ||
| 42 | MAP2K1 | 12474 | -0.704 | -0.4731 | No | ||
| 43 | IFNA4 | 12517 | -0.717 | -0.4732 | No | ||
| 44 | KLRC3 | 12606 | -0.746 | -0.4758 | No | ||
| 45 | IFNAR1 | 12692 | -0.777 | -0.4780 | No | ||
| 46 | SOS1 | 12695 | -0.778 | -0.4758 | No | ||
| 47 | PIK3R1 | 12882 | -0.833 | -0.4834 | No | ||
| 48 | ITGAL | 13227 | -0.958 | -0.4991 | No | ||
| 49 | GZMB | 13508 | -1.072 | -0.5110 | No | ||
| 50 | PLCG1 | 13591 | -1.102 | -0.5122 | No | ||
| 51 | CSF2 | 13610 | -1.107 | -0.5099 | No | ||
| 52 | RAC1 | 13719 | -1.142 | -0.5123 | No | ||
| 53 | TNF | 13737 | -1.150 | -0.5098 | No | ||
| 54 | KLRC2 | 13798 | -1.181 | -0.5095 | No | ||
| 55 | KLRD1 | 14069 | -1.307 | -0.5202 | No | ||
| 56 | ARAF | 14425 | -1.487 | -0.5349 | No | ||
| 57 | FASLG | 14448 | -1.502 | -0.5317 | No | ||
| 58 | ZAP70 | 14876 | -1.733 | -0.5496 | No | ||
| 59 | PPP3CA | 14970 | -1.793 | -0.5492 | No | ||
| 60 | IFNA5 | 14974 | -1.794 | -0.5441 | No | ||
| 61 | IFNGR1 | 15024 | -1.836 | -0.5412 | No | ||
| 62 | LCK | 15211 | -1.955 | -0.5455 | No | ||
| 63 | FYN | 15378 | -2.088 | -0.5482 | No | ||
| 64 | PIK3CA | 15572 | -2.276 | -0.5519 | No | ||
| 65 | VAV2 | 15699 | -2.400 | -0.5515 | No | ||
| 66 | IFNGR2 | 16004 | -2.747 | -0.5598 | Yes | ||
| 67 | NFAT5 | 16018 | -2.763 | -0.5522 | Yes | ||
| 68 | PPP3R1 | 16123 | -2.886 | -0.5493 | Yes | ||
| 69 | RAF1 | 16233 | -3.067 | -0.5460 | Yes | ||
| 70 | ICAM1 | 16314 | -3.203 | -0.5408 | Yes | ||
| 71 | PPP3CB | 16355 | -3.264 | -0.5332 | Yes | ||
| 72 | PPP3CC | 16365 | -3.277 | -0.5240 | Yes | ||
| 73 | NFATC3 | 16616 | -3.788 | -0.5262 | Yes | ||
| 74 | PIK3CD | 16811 | -4.218 | -0.5241 | Yes | ||
| 75 | ITGB2 | 16835 | -4.266 | -0.5127 | Yes | ||
| 76 | IFNG | 16860 | -4.309 | -0.5011 | Yes | ||
| 77 | SOS2 | 17023 | -4.655 | -0.4960 | Yes | ||
| 78 | FAS | 17062 | -4.760 | -0.4839 | Yes | ||
| 79 | SH3BP2 | 17072 | -4.786 | -0.4701 | Yes | ||
| 80 | MAPK1 | 17264 | -5.380 | -0.4644 | Yes | ||
| 81 | GRB2 | 17489 | -6.003 | -0.4587 | Yes | ||
| 82 | MAPK3 | 17495 | -6.022 | -0.4410 | Yes | ||
| 83 | PRKCB1 | 17557 | -6.270 | -0.4256 | Yes | ||
| 84 | PLCG2 | 17677 | -6.671 | -0.4122 | Yes | ||
| 85 | ICAM2 | 17999 | -8.264 | -0.4049 | Yes | ||
| 86 | PRKCA | 18108 | -9.028 | -0.3839 | Yes | ||
| 87 | IFNAR2 | 18260 | -10.414 | -0.3610 | Yes | ||
| 88 | VAV1 | 18300 | -10.774 | -0.3311 | Yes | ||
| 89 | PIK3CG | 18341 | -11.438 | -0.2992 | Yes | ||
| 90 | SYK | 18377 | -12.306 | -0.2644 | Yes | ||
| 91 | KRAS | 18389 | -12.525 | -0.2277 | Yes | ||
| 92 | NFATC1 | 18423 | -13.291 | -0.1900 | Yes | ||
| 93 | RAC2 | 18482 | -15.379 | -0.1473 | Yes | ||
| 94 | PTK2B | 18488 | -15.600 | -0.1011 | Yes | ||
| 95 | PTPN6 | 18504 | -16.488 | -0.0529 | Yes | ||
| 96 | VAV3 | 18534 | -19.775 | 0.0044 | Yes |