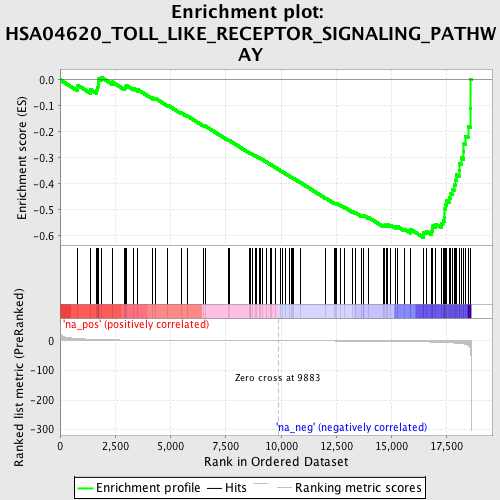
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04620_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.6065689 |
| Normalized Enrichment Score (NES) | -1.822464 |
| Nominal p-value | 0.014842301 |
| FDR q-value | 0.35300595 |
| FWER p-Value | 0.932 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PIK3CB | 780 | 7.582 | -0.0203 | No | ||
| 2 | MAPK12 | 1388 | 5.094 | -0.0384 | No | ||
| 3 | PIK3R3 | 1649 | 4.388 | -0.0398 | No | ||
| 4 | MAPK8 | 1673 | 4.351 | -0.0285 | No | ||
| 5 | MAP3K7 | 1717 | 4.246 | -0.0185 | No | ||
| 6 | MAPK11 | 1722 | 4.235 | -0.0066 | No | ||
| 7 | TICAM2 | 1726 | 4.224 | 0.0054 | No | ||
| 8 | CD86 | 1882 | 3.880 | 0.0083 | No | ||
| 9 | MYD88 | 2356 | 3.069 | -0.0084 | No | ||
| 10 | CXCL9 | 2926 | 2.453 | -0.0321 | No | ||
| 11 | AKT1 | 2954 | 2.432 | -0.0265 | No | ||
| 12 | TIRAP | 3007 | 2.381 | -0.0225 | No | ||
| 13 | IFNA1 | 3323 | 2.127 | -0.0334 | No | ||
| 14 | CHUK | 3524 | 1.992 | -0.0384 | No | ||
| 15 | IFNA7 | 4188 | 1.647 | -0.0695 | No | ||
| 16 | IRAK1 | 4314 | 1.586 | -0.0717 | No | ||
| 17 | TLR3 | 4876 | 1.365 | -0.0980 | No | ||
| 18 | IKBKB | 5479 | 1.167 | -0.1272 | No | ||
| 19 | IFNA2 | 5761 | 1.074 | -0.1392 | No | ||
| 20 | CD14 | 6488 | 0.857 | -0.1760 | No | ||
| 21 | CCL4 | 6567 | 0.833 | -0.1778 | No | ||
| 22 | PIK3R2 | 7604 | 0.563 | -0.2321 | No | ||
| 23 | IFNB1 | 7668 | 0.547 | -0.2339 | No | ||
| 24 | IFNA6 | 8566 | 0.320 | -0.2814 | No | ||
| 25 | TRAF3 | 8599 | 0.313 | -0.2822 | No | ||
| 26 | TLR5 | 8633 | 0.305 | -0.2831 | No | ||
| 27 | MAP2K7 | 8699 | 0.290 | -0.2858 | No | ||
| 28 | IFNA14 | 8836 | 0.256 | -0.2924 | No | ||
| 29 | PIK3R5 | 8866 | 0.250 | -0.2933 | No | ||
| 30 | IL1B | 9018 | 0.212 | -0.3008 | No | ||
| 31 | MAPK13 | 9080 | 0.198 | -0.3035 | No | ||
| 32 | TLR7 | 9146 | 0.184 | -0.3065 | No | ||
| 33 | CXCL10 | 9329 | 0.138 | -0.3159 | No | ||
| 34 | CCL3 | 9544 | 0.084 | -0.3272 | No | ||
| 35 | CCL5 | 9577 | 0.075 | -0.3288 | No | ||
| 36 | LBP | 9762 | 0.033 | -0.3386 | No | ||
| 37 | NFKB2 | 9984 | -0.028 | -0.3505 | No | ||
| 38 | IKBKE | 10076 | -0.051 | -0.3552 | No | ||
| 39 | CXCL11 | 10204 | -0.079 | -0.3618 | No | ||
| 40 | TLR8 | 10360 | -0.119 | -0.3699 | No | ||
| 41 | FOS | 10490 | -0.151 | -0.3764 | No | ||
| 42 | IL6 | 10537 | -0.164 | -0.3784 | No | ||
| 43 | IL12B | 10559 | -0.169 | -0.3791 | No | ||
| 44 | CD80 | 10879 | -0.257 | -0.3955 | No | ||
| 45 | IFNA13 | 12022 | -0.564 | -0.4556 | No | ||
| 46 | MAP2K2 | 12431 | -0.690 | -0.4756 | No | ||
| 47 | MAP2K1 | 12474 | -0.704 | -0.4759 | No | ||
| 48 | IFNA4 | 12517 | -0.717 | -0.4761 | No | ||
| 49 | IFNAR1 | 12692 | -0.777 | -0.4832 | No | ||
| 50 | PIK3R1 | 12882 | -0.833 | -0.4910 | No | ||
| 51 | MAPK10 | 13249 | -0.964 | -0.5080 | No | ||
| 52 | IRF7 | 13372 | -1.008 | -0.5117 | No | ||
| 53 | RIPK1 | 13643 | -1.118 | -0.5230 | No | ||
| 54 | RAC1 | 13719 | -1.142 | -0.5238 | No | ||
| 55 | TNF | 13737 | -1.150 | -0.5214 | No | ||
| 56 | MAP3K8 | 13974 | -1.256 | -0.5305 | No | ||
| 57 | TLR4 | 14638 | -1.600 | -0.5617 | No | ||
| 58 | FADD | 14667 | -1.621 | -0.5585 | No | ||
| 59 | LY96 | 14777 | -1.674 | -0.5596 | No | ||
| 60 | TLR9 | 14809 | -1.692 | -0.5564 | No | ||
| 61 | IFNA5 | 14974 | -1.794 | -0.5601 | No | ||
| 62 | TBK1 | 15179 | -1.931 | -0.5655 | No | ||
| 63 | MAP3K7IP1 | 15267 | -1.988 | -0.5645 | No | ||
| 64 | PIK3CA | 15572 | -2.276 | -0.5744 | No | ||
| 65 | IRF5 | 15854 | -2.562 | -0.5822 | No | ||
| 66 | MAP2K3 | 15856 | -2.564 | -0.5748 | No | ||
| 67 | MAP2K4 | 16445 | -3.446 | -0.5966 | Yes | ||
| 68 | IKBKG | 16460 | -3.469 | -0.5874 | Yes | ||
| 69 | CASP8 | 16569 | -3.698 | -0.5826 | Yes | ||
| 70 | PIK3CD | 16811 | -4.218 | -0.5834 | Yes | ||
| 71 | JUN | 16845 | -4.286 | -0.5729 | Yes | ||
| 72 | MAPK14 | 16874 | -4.333 | -0.5619 | Yes | ||
| 73 | MAP2K6 | 16991 | -4.589 | -0.5549 | Yes | ||
| 74 | MAPK1 | 17264 | -5.380 | -0.5541 | Yes | ||
| 75 | IRAK4 | 17330 | -5.552 | -0.5416 | Yes | ||
| 76 | NFKB1 | 17383 | -5.733 | -0.5279 | Yes | ||
| 77 | IL12A | 17415 | -5.813 | -0.5128 | Yes | ||
| 78 | RELA | 17416 | -5.813 | -0.4961 | Yes | ||
| 79 | AKT3 | 17454 | -5.931 | -0.4810 | Yes | ||
| 80 | MAPK3 | 17495 | -6.022 | -0.4658 | Yes | ||
| 81 | MAPK9 | 17613 | -6.461 | -0.4535 | Yes | ||
| 82 | TOLLIP | 17688 | -6.703 | -0.4382 | Yes | ||
| 83 | IRF3 | 17778 | -7.066 | -0.4226 | Yes | ||
| 84 | AKT2 | 17832 | -7.269 | -0.4046 | Yes | ||
| 85 | NFKBIA | 17884 | -7.579 | -0.3855 | Yes | ||
| 86 | CD40 | 17926 | -7.796 | -0.3652 | Yes | ||
| 87 | MAP3K7IP2 | 18062 | -8.635 | -0.3476 | Yes | ||
| 88 | TRAF6 | 18077 | -8.765 | -0.3231 | Yes | ||
| 89 | TLR2 | 18156 | -9.445 | -0.3001 | Yes | ||
| 90 | IFNAR2 | 18260 | -10.414 | -0.2757 | Yes | ||
| 91 | STAT1 | 18276 | -10.570 | -0.2461 | Yes | ||
| 92 | PIK3CG | 18341 | -11.438 | -0.2166 | Yes | ||
| 93 | TLR1 | 18467 | -14.848 | -0.1805 | Yes | ||
| 94 | SPP1 | 18561 | -25.847 | -0.1111 | Yes | ||
| 95 | TLR6 | 18585 | -39.562 | 0.0017 | Yes |
