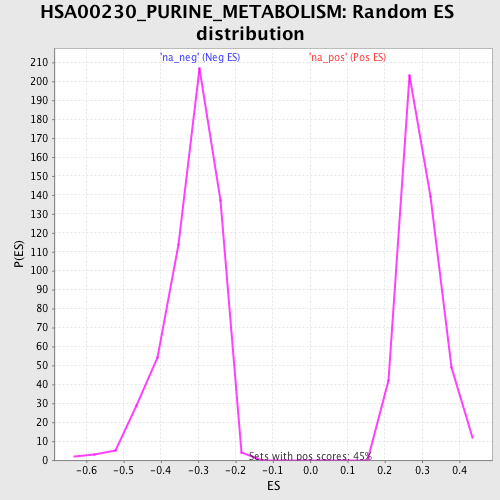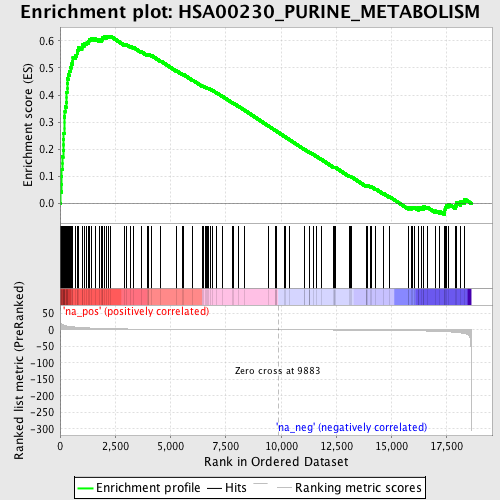
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA00230_PURINE_METABOLISM |
| Enrichment Score (ES) | 0.617162 |
| Normalized Enrichment Score (NES) | 2.094684 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | POLR1B | 14 | 28.191 | 0.0445 | Yes | ||
| 2 | IMPDH2 | 58 | 18.288 | 0.0715 | Yes | ||
| 3 | NME2 | 66 | 17.853 | 0.0997 | Yes | ||
| 4 | ADK | 79 | 16.597 | 0.1257 | Yes | ||
| 5 | NME1 | 107 | 15.161 | 0.1485 | Yes | ||
| 6 | PPAT | 110 | 15.096 | 0.1727 | Yes | ||
| 7 | POLD2 | 133 | 14.463 | 0.1947 | Yes | ||
| 8 | GART | 150 | 13.835 | 0.2160 | Yes | ||
| 9 | AK2 | 153 | 13.651 | 0.2378 | Yes | ||
| 10 | APRT | 165 | 13.403 | 0.2587 | Yes | ||
| 11 | ADSL | 186 | 12.945 | 0.2784 | Yes | ||
| 12 | POLR3H | 187 | 12.912 | 0.2991 | Yes | ||
| 13 | NME6 | 188 | 12.892 | 0.3197 | Yes | ||
| 14 | ADA | 218 | 12.375 | 0.3380 | Yes | ||
| 15 | POLR1A | 229 | 12.237 | 0.3571 | Yes | ||
| 16 | NME4 | 274 | 11.661 | 0.3734 | Yes | ||
| 17 | GMPS | 291 | 11.420 | 0.3909 | Yes | ||
| 18 | PKM2 | 293 | 11.356 | 0.4090 | Yes | ||
| 19 | POLE3 | 316 | 11.119 | 0.4257 | Yes | ||
| 20 | POLE | 321 | 11.089 | 0.4433 | Yes | ||
| 21 | NUDT5 | 337 | 10.876 | 0.4599 | Yes | ||
| 22 | ATIC | 370 | 10.520 | 0.4750 | Yes | ||
| 23 | POLR2J | 428 | 9.986 | 0.4880 | Yes | ||
| 24 | POLR2H | 482 | 9.582 | 0.5005 | Yes | ||
| 25 | GUK1 | 500 | 9.459 | 0.5147 | Yes | ||
| 26 | RFC5 | 570 | 8.951 | 0.5253 | Yes | ||
| 27 | NUDT9 | 580 | 8.858 | 0.5391 | Yes | ||
| 28 | ENTPD4 | 691 | 8.119 | 0.5461 | Yes | ||
| 29 | PAICS | 764 | 7.678 | 0.5546 | Yes | ||
| 30 | AMPD2 | 789 | 7.536 | 0.5653 | Yes | ||
| 31 | NUDT2 | 821 | 7.395 | 0.5755 | Yes | ||
| 32 | NME7 | 991 | 6.502 | 0.5768 | Yes | ||
| 33 | POLA2 | 997 | 6.482 | 0.5869 | Yes | ||
| 34 | POLR2E | 1117 | 5.985 | 0.5901 | Yes | ||
| 35 | HPRT1 | 1215 | 5.665 | 0.5939 | Yes | ||
| 36 | POLR2K | 1286 | 5.454 | 0.5989 | Yes | ||
| 37 | RRM1 | 1310 | 5.381 | 0.6063 | Yes | ||
| 38 | PRPS1 | 1398 | 5.067 | 0.6097 | Yes | ||
| 39 | POLR1D | 1584 | 4.549 | 0.6070 | Yes | ||
| 40 | POLR3A | 1767 | 4.133 | 0.6038 | Yes | ||
| 41 | POLR2A | 1865 | 3.916 | 0.6048 | Yes | ||
| 42 | PRIM1 | 1905 | 3.839 | 0.6089 | Yes | ||
| 43 | POLD3 | 1927 | 3.807 | 0.6138 | Yes | ||
| 44 | PDE8A | 2020 | 3.635 | 0.6147 | Yes | ||
| 45 | PNPT1 | 2111 | 3.476 | 0.6154 | Yes | ||
| 46 | POLR2I | 2179 | 3.359 | 0.6172 | Yes | ||
| 47 | NT5E | 2287 | 3.179 | 0.6165 | No | ||
| 48 | AK3L1 | 2925 | 2.453 | 0.5860 | No | ||
| 49 | ENTPD6 | 2988 | 2.398 | 0.5864 | No | ||
| 50 | ECGF1 | 3175 | 2.229 | 0.5800 | No | ||
| 51 | PRPS2 | 3314 | 2.133 | 0.5759 | No | ||
| 52 | ADCY3 | 3667 | 1.902 | 0.5599 | No | ||
| 53 | AK5 | 3947 | 1.764 | 0.5477 | No | ||
| 54 | PDE10A | 4002 | 1.738 | 0.5475 | No | ||
| 55 | POLR3B | 4006 | 1.736 | 0.5502 | No | ||
| 56 | ENTPD1 | 4154 | 1.666 | 0.5449 | No | ||
| 57 | PDE6G | 4562 | 1.485 | 0.5252 | No | ||
| 58 | NT5C3 | 5262 | 1.236 | 0.4894 | No | ||
| 59 | NP | 5532 | 1.149 | 0.4767 | No | ||
| 60 | POLR2G | 5592 | 1.130 | 0.4753 | No | ||
| 61 | ENTPD8 | 6000 | 1.007 | 0.4549 | No | ||
| 62 | GUCY2C | 6424 | 0.875 | 0.4335 | No | ||
| 63 | ADCY4 | 6491 | 0.857 | 0.4313 | No | ||
| 64 | PDE8B | 6560 | 0.835 | 0.4289 | No | ||
| 65 | IMPDH1 | 6608 | 0.822 | 0.4277 | No | ||
| 66 | AMPD3 | 6676 | 0.803 | 0.4254 | No | ||
| 67 | ALLC | 6730 | 0.792 | 0.4238 | No | ||
| 68 | RRM2B | 6818 | 0.770 | 0.4203 | No | ||
| 69 | GUCY1A3 | 6903 | 0.750 | 0.4170 | No | ||
| 70 | ENTPD3 | 7090 | 0.700 | 0.4080 | No | ||
| 71 | GUCY2F | 7328 | 0.635 | 0.3962 | No | ||
| 72 | GUCY1A2 | 7796 | 0.514 | 0.3718 | No | ||
| 73 | POLR3K | 7847 | 0.497 | 0.3699 | No | ||
| 74 | PDE5A | 8051 | 0.447 | 0.3596 | No | ||
| 75 | AK1 | 8345 | 0.376 | 0.3444 | No | ||
| 76 | NPR1 | 9442 | 0.105 | 0.2853 | No | ||
| 77 | XDH | 9736 | 0.041 | 0.2695 | No | ||
| 78 | POLA1 | 9763 | 0.033 | 0.2681 | No | ||
| 79 | GDA | 9794 | 0.025 | 0.2665 | No | ||
| 80 | GUCY1B3 | 9806 | 0.023 | 0.2660 | No | ||
| 81 | PDE9A | 10135 | -0.065 | 0.2483 | No | ||
| 82 | ITPA | 10186 | -0.075 | 0.2458 | No | ||
| 83 | ENPP1 | 10385 | -0.125 | 0.2352 | No | ||
| 84 | PDE3B | 11038 | -0.299 | 0.2005 | No | ||
| 85 | ADCY8 | 11073 | -0.313 | 0.1991 | No | ||
| 86 | POLE2 | 11285 | -0.370 | 0.1883 | No | ||
| 87 | ADCY2 | 11292 | -0.372 | 0.1886 | No | ||
| 88 | PDE1A | 11296 | -0.373 | 0.1890 | No | ||
| 89 | PDE2A | 11481 | -0.421 | 0.1797 | No | ||
| 90 | AMPD1 | 11616 | -0.461 | 0.1732 | No | ||
| 91 | ENPP3 | 11818 | -0.509 | 0.1632 | No | ||
| 92 | PKLR | 12376 | -0.675 | 0.1341 | No | ||
| 93 | PAPSS2 | 12432 | -0.691 | 0.1323 | No | ||
| 94 | PDE1C | 12440 | -0.694 | 0.1330 | No | ||
| 95 | ADCY5 | 12461 | -0.699 | 0.1330 | No | ||
| 96 | PDE7B | 13098 | -0.911 | 0.1001 | No | ||
| 97 | POLR2C | 13132 | -0.928 | 0.0998 | No | ||
| 98 | ADCY6 | 13195 | -0.946 | 0.0980 | No | ||
| 99 | PDE4B | 13854 | -1.203 | 0.0643 | No | ||
| 100 | POLR2B | 13857 | -1.203 | 0.0661 | No | ||
| 101 | PDE6H | 13866 | -1.205 | 0.0676 | No | ||
| 102 | ENTPD2 | 13926 | -1.234 | 0.0664 | No | ||
| 103 | FHIT | 14059 | -1.302 | 0.0614 | No | ||
| 104 | CANT1 | 14089 | -1.313 | 0.0619 | No | ||
| 105 | NPR2 | 14283 | -1.411 | 0.0537 | No | ||
| 106 | NT5C1B | 14636 | -1.600 | 0.0372 | No | ||
| 107 | PDE11A | 14911 | -1.752 | 0.0252 | No | ||
| 108 | PDE4D | 15754 | -2.452 | -0.0164 | No | ||
| 109 | PAPSS1 | 15778 | -2.478 | -0.0136 | No | ||
| 110 | NT5C | 15902 | -2.618 | -0.0161 | No | ||
| 111 | GMPR | 15933 | -2.663 | -0.0135 | No | ||
| 112 | POLE4 | 16044 | -2.798 | -0.0149 | No | ||
| 113 | ZNRD1 | 16215 | -3.030 | -0.0193 | No | ||
| 114 | GMPR2 | 16223 | -3.045 | -0.0147 | No | ||
| 115 | PDE4C | 16337 | -3.245 | -0.0157 | No | ||
| 116 | ADCY9 | 16439 | -3.439 | -0.0156 | No | ||
| 117 | PDE4A | 16459 | -3.468 | -0.0111 | No | ||
| 118 | NT5M | 16618 | -3.790 | -0.0135 | No | ||
| 119 | NT5C2 | 17007 | -4.626 | -0.0271 | No | ||
| 120 | ENTPD5 | 17180 | -5.116 | -0.0282 | No | ||
| 121 | RRM2 | 17403 | -5.780 | -0.0309 | No | ||
| 122 | ADCY7 | 17418 | -5.824 | -0.0224 | No | ||
| 123 | ADSSL1 | 17431 | -5.873 | -0.0136 | No | ||
| 124 | POLD1 | 17475 | -5.968 | -0.0063 | No | ||
| 125 | DCK | 17594 | -6.389 | -0.0025 | No | ||
| 126 | PDE7A | 17899 | -7.661 | -0.0066 | No | ||
| 127 | DGUOK | 17948 | -7.939 | 0.0035 | No | ||
| 128 | PDE6D | 18136 | -9.289 | 0.0083 | No | ||
| 129 | POLD4 | 18311 | -10.982 | 0.0165 | No |