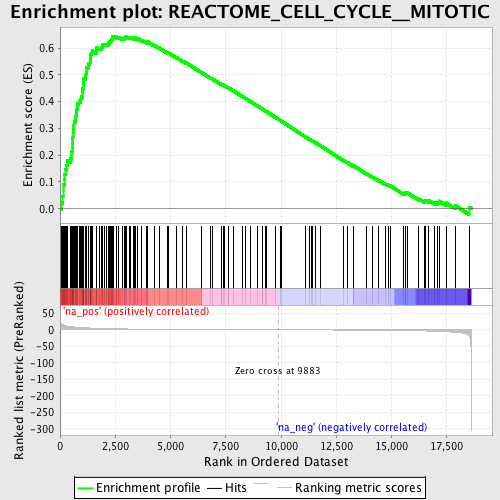
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.64482415 |
| Normalized Enrichment Score (NES) | 2.2294853 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 76 | 16.722 | 0.0233 | Yes | ||
| 2 | NUP43 | 121 | 14.890 | 0.0453 | Yes | ||
| 3 | POLD2 | 133 | 14.463 | 0.0684 | Yes | ||
| 4 | CDK4 | 134 | 14.452 | 0.0921 | Yes | ||
| 5 | PSMD8 | 200 | 12.625 | 0.1092 | Yes | ||
| 6 | CCNE1 | 210 | 12.512 | 0.1293 | Yes | ||
| 7 | PSMA7 | 233 | 12.156 | 0.1480 | Yes | ||
| 8 | CDC16 | 301 | 11.280 | 0.1628 | Yes | ||
| 9 | POLE | 321 | 11.089 | 0.1800 | Yes | ||
| 10 | PSMC5 | 473 | 9.639 | 0.1876 | Yes | ||
| 11 | ORC1L | 524 | 9.323 | 0.2002 | Yes | ||
| 12 | RPA2 | 529 | 9.268 | 0.2151 | Yes | ||
| 13 | TUBG1 | 553 | 9.071 | 0.2288 | Yes | ||
| 14 | NUP37 | 557 | 9.038 | 0.2434 | Yes | ||
| 15 | RFC5 | 570 | 8.951 | 0.2574 | Yes | ||
| 16 | PSMD2 | 581 | 8.853 | 0.2714 | Yes | ||
| 17 | NUP107 | 592 | 8.768 | 0.2852 | Yes | ||
| 18 | TYMS | 615 | 8.624 | 0.2981 | Yes | ||
| 19 | MCM7 | 624 | 8.576 | 0.3118 | Yes | ||
| 20 | PSMD7 | 628 | 8.552 | 0.3256 | Yes | ||
| 21 | UBE2E1 | 687 | 8.131 | 0.3358 | Yes | ||
| 22 | RFC4 | 717 | 7.960 | 0.3473 | Yes | ||
| 23 | ORC2L | 724 | 7.897 | 0.3599 | Yes | ||
| 24 | RPA1 | 738 | 7.796 | 0.3720 | Yes | ||
| 25 | RFC3 | 771 | 7.657 | 0.3828 | Yes | ||
| 26 | MCM3 | 785 | 7.561 | 0.3944 | Yes | ||
| 27 | ORC5L | 854 | 7.204 | 0.4026 | Yes | ||
| 28 | MCM10 | 940 | 6.780 | 0.4091 | Yes | ||
| 29 | CDC25A | 963 | 6.668 | 0.4188 | Yes | ||
| 30 | FGFR1OP | 996 | 6.489 | 0.4277 | Yes | ||
| 31 | POLA2 | 997 | 6.482 | 0.4383 | Yes | ||
| 32 | PSMD9 | 1022 | 6.383 | 0.4475 | Yes | ||
| 33 | PSMD3 | 1038 | 6.338 | 0.4571 | Yes | ||
| 34 | LIG1 | 1042 | 6.323 | 0.4673 | Yes | ||
| 35 | MCM4 | 1059 | 6.251 | 0.4766 | Yes | ||
| 36 | PSMB2 | 1066 | 6.195 | 0.4865 | Yes | ||
| 37 | ZWINT | 1143 | 5.913 | 0.4920 | Yes | ||
| 38 | BUB1B | 1156 | 5.882 | 0.5010 | Yes | ||
| 39 | CDC45L | 1175 | 5.835 | 0.5096 | Yes | ||
| 40 | MCM5 | 1196 | 5.762 | 0.5180 | Yes | ||
| 41 | PSMA4 | 1197 | 5.761 | 0.5274 | Yes | ||
| 42 | MCM2 | 1266 | 5.511 | 0.5328 | Yes | ||
| 43 | GMNN | 1272 | 5.493 | 0.5415 | Yes | ||
| 44 | FEN1 | 1352 | 5.246 | 0.5458 | Yes | ||
| 45 | PSME3 | 1362 | 5.209 | 0.5539 | Yes | ||
| 46 | RFC1 | 1375 | 5.143 | 0.5616 | Yes | ||
| 47 | KNTC1 | 1386 | 5.099 | 0.5694 | Yes | ||
| 48 | PRKAR2B | 1389 | 5.093 | 0.5777 | Yes | ||
| 49 | PSMD12 | 1419 | 4.991 | 0.5843 | Yes | ||
| 50 | PSMC4 | 1469 | 4.838 | 0.5896 | Yes | ||
| 51 | PSMB5 | 1637 | 4.411 | 0.5878 | Yes | ||
| 52 | RPA3 | 1650 | 4.387 | 0.5943 | Yes | ||
| 53 | PSMA1 | 1655 | 4.376 | 0.6012 | Yes | ||
| 54 | CDK2 | 1797 | 4.060 | 0.6003 | Yes | ||
| 55 | ZW10 | 1874 | 3.892 | 0.6025 | Yes | ||
| 56 | PRIM1 | 1905 | 3.839 | 0.6072 | Yes | ||
| 57 | POLD3 | 1927 | 3.807 | 0.6123 | Yes | ||
| 58 | PSMC6 | 1990 | 3.688 | 0.6150 | Yes | ||
| 59 | DCTN2 | 2116 | 3.465 | 0.6139 | Yes | ||
| 60 | CUL1 | 2177 | 3.362 | 0.6162 | Yes | ||
| 61 | RANBP2 | 2207 | 3.299 | 0.6200 | Yes | ||
| 62 | BIRC5 | 2238 | 3.261 | 0.6237 | Yes | ||
| 63 | PSMD14 | 2294 | 3.168 | 0.6259 | Yes | ||
| 64 | PSMB4 | 2328 | 3.116 | 0.6292 | Yes | ||
| 65 | CENPE | 2339 | 3.102 | 0.6338 | Yes | ||
| 66 | CCNH | 2354 | 3.077 | 0.6381 | Yes | ||
| 67 | NUP160 | 2376 | 3.042 | 0.6419 | Yes | ||
| 68 | TUBA1A | 2414 | 2.999 | 0.6448 | Yes | ||
| 69 | YWHAE | 2553 | 2.833 | 0.6420 | No | ||
| 70 | PSMA5 | 2658 | 2.718 | 0.6408 | No | ||
| 71 | CCNB1 | 2806 | 2.558 | 0.6371 | No | ||
| 72 | PCNA | 2909 | 2.465 | 0.6356 | No | ||
| 73 | PSMC1 | 2914 | 2.462 | 0.6394 | No | ||
| 74 | MNAT1 | 2949 | 2.434 | 0.6415 | No | ||
| 75 | PSMD11 | 2983 | 2.403 | 0.6437 | No | ||
| 76 | WEE1 | 3159 | 2.243 | 0.6379 | No | ||
| 77 | PRKACA | 3166 | 2.234 | 0.6412 | No | ||
| 78 | PSMA2 | 3324 | 2.126 | 0.6362 | No | ||
| 79 | DHFR | 3368 | 2.094 | 0.6373 | No | ||
| 80 | PSMC3 | 3404 | 2.066 | 0.6388 | No | ||
| 81 | CLIP1 | 3518 | 1.997 | 0.6360 | No | ||
| 82 | SGOL1 | 3700 | 1.888 | 0.6293 | No | ||
| 83 | SDCCAG8 | 3911 | 1.777 | 0.6208 | No | ||
| 84 | PSMA3 | 3935 | 1.768 | 0.6225 | No | ||
| 85 | KIF20A | 3966 | 1.754 | 0.6237 | No | ||
| 86 | SMC1A | 4261 | 1.606 | 0.6104 | No | ||
| 87 | CDC25C | 4477 | 1.518 | 0.6013 | No | ||
| 88 | PLK4 | 4862 | 1.371 | 0.5827 | No | ||
| 89 | CCNA2 | 4888 | 1.360 | 0.5836 | No | ||
| 90 | TUBA4A | 5274 | 1.232 | 0.5648 | No | ||
| 91 | KIF23 | 5551 | 1.142 | 0.5517 | No | ||
| 92 | RAD21 | 5708 | 1.092 | 0.5451 | No | ||
| 93 | XPO1 | 6402 | 0.881 | 0.5090 | No | ||
| 94 | SEC13 | 6820 | 0.769 | 0.4877 | No | ||
| 95 | BUB1 | 6907 | 0.749 | 0.4842 | No | ||
| 96 | PSMD5 | 7292 | 0.647 | 0.4645 | No | ||
| 97 | DYNLL1 | 7294 | 0.646 | 0.4655 | No | ||
| 98 | RANGAP1 | 7372 | 0.623 | 0.4624 | No | ||
| 99 | PLK1 | 7456 | 0.602 | 0.4589 | No | ||
| 100 | PSMA6 | 7598 | 0.565 | 0.4522 | No | ||
| 101 | OFD1 | 7612 | 0.562 | 0.4524 | No | ||
| 102 | DCTN3 | 7866 | 0.492 | 0.4395 | No | ||
| 103 | TUBB4 | 8257 | 0.396 | 0.4190 | No | ||
| 104 | PSMD4 | 8384 | 0.365 | 0.4128 | No | ||
| 105 | NEK2 | 8608 | 0.311 | 0.4012 | No | ||
| 106 | PSMB1 | 8935 | 0.231 | 0.3840 | No | ||
| 107 | PMF1 | 9169 | 0.178 | 0.3717 | No | ||
| 108 | E2F1 | 9178 | 0.176 | 0.3715 | No | ||
| 109 | CCNA1 | 9282 | 0.148 | 0.3662 | No | ||
| 110 | CCNB2 | 9319 | 0.141 | 0.3645 | No | ||
| 111 | POLA1 | 9763 | 0.033 | 0.3405 | No | ||
| 112 | CDK7 | 9958 | -0.022 | 0.3301 | No | ||
| 113 | NDC80 | 10000 | -0.032 | 0.3279 | No | ||
| 114 | NUMA1 | 11085 | -0.317 | 0.2697 | No | ||
| 115 | POLE2 | 11285 | -0.370 | 0.2596 | No | ||
| 116 | PPP1CC | 11390 | -0.396 | 0.2546 | No | ||
| 117 | MAD2L1 | 11436 | -0.406 | 0.2528 | No | ||
| 118 | KIF2A | 11577 | -0.447 | 0.2460 | No | ||
| 119 | BUB3 | 11776 | -0.499 | 0.2361 | No | ||
| 120 | SSNA1 | 12848 | -0.823 | 0.1795 | No | ||
| 121 | CDC25B | 12990 | -0.869 | 0.1733 | No | ||
| 122 | CLASP1 | 13281 | -0.976 | 0.1592 | No | ||
| 123 | CETN2 | 13295 | -0.979 | 0.1601 | No | ||
| 124 | UBE2D1 | 13861 | -1.204 | 0.1314 | No | ||
| 125 | STAG2 | 14159 | -1.349 | 0.1176 | No | ||
| 126 | UBE2C | 14431 | -1.492 | 0.1054 | No | ||
| 127 | PTTG1 | 14724 | -1.650 | 0.0923 | No | ||
| 128 | CLASP2 | 14840 | -1.710 | 0.0888 | No | ||
| 129 | CENPC1 | 14952 | -1.781 | 0.0857 | No | ||
| 130 | YWHAG | 15545 | -2.257 | 0.0574 | No | ||
| 131 | PAFAH1B1 | 15623 | -2.326 | 0.0570 | No | ||
| 132 | ORC4L | 15649 | -2.357 | 0.0595 | No | ||
| 133 | SGOL2 | 15709 | -2.412 | 0.0603 | No | ||
| 134 | CDC20 | 16222 | -3.043 | 0.0376 | No | ||
| 135 | PSMB10 | 16501 | -3.561 | 0.0284 | No | ||
| 136 | RB1 | 16538 | -3.642 | 0.0324 | No | ||
| 137 | MAPRE1 | 16691 | -3.930 | 0.0306 | No | ||
| 138 | CSNK1E | 16965 | -4.536 | 0.0232 | No | ||
| 139 | CENPA | 17099 | -4.886 | 0.0240 | No | ||
| 140 | ACTR1A | 17162 | -5.066 | 0.0290 | No | ||
| 141 | POLD1 | 17475 | -5.968 | 0.0219 | No | ||
| 142 | CDKN1A | 17893 | -7.620 | 0.0118 | No | ||
| 143 | TK2 | 18507 | -16.654 | 0.0059 | No |