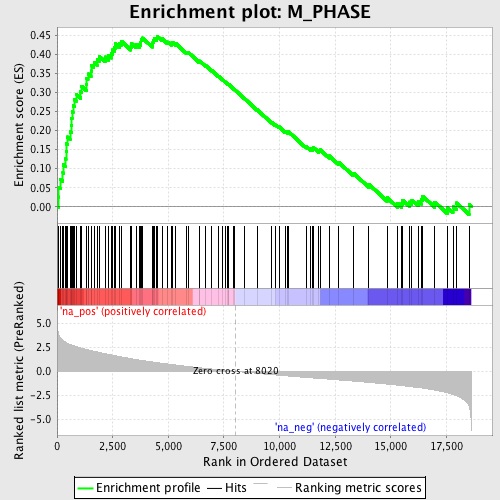
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | M_PHASE |
| Enrichment Score (ES) | 0.44784552 |
| Normalized Enrichment Score (NES) | 2.0808713 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.007514629 |
| FWER p-Value | 0.069 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TGFA | 49 | 4.065 | 0.0258 | Yes | ||
| 2 | ANLN | 70 | 3.873 | 0.0519 | Yes | ||
| 3 | NDC80 | 138 | 3.521 | 0.0729 | Yes | ||
| 4 | NCAPH | 260 | 3.228 | 0.0890 | Yes | ||
| 5 | SMC1A | 277 | 3.191 | 0.1105 | Yes | ||
| 6 | CCNA2 | 386 | 3.023 | 0.1258 | Yes | ||
| 7 | KIF15 | 401 | 2.995 | 0.1460 | Yes | ||
| 8 | RAD50 | 422 | 2.966 | 0.1657 | Yes | ||
| 9 | TPX2 | 474 | 2.908 | 0.1833 | Yes | ||
| 10 | EGF | 585 | 2.793 | 0.1969 | Yes | ||
| 11 | NUSAP1 | 636 | 2.749 | 0.2135 | Yes | ||
| 12 | SPO11 | 655 | 2.732 | 0.2317 | Yes | ||
| 13 | AKAP8 | 674 | 2.714 | 0.2497 | Yes | ||
| 14 | MAD2L1 | 718 | 2.685 | 0.2662 | Yes | ||
| 15 | BUB1B | 774 | 2.637 | 0.2817 | Yes | ||
| 16 | KPNA2 | 855 | 2.577 | 0.2954 | Yes | ||
| 17 | SYCP1 | 1056 | 2.444 | 0.3017 | Yes | ||
| 18 | ANAPC4 | 1093 | 2.423 | 0.3168 | Yes | ||
| 19 | RAD51L1 | 1316 | 2.290 | 0.3208 | Yes | ||
| 20 | SMC3 | 1328 | 2.281 | 0.3362 | Yes | ||
| 21 | NBN | 1390 | 2.246 | 0.3486 | Yes | ||
| 22 | RAD21 | 1527 | 2.167 | 0.3565 | Yes | ||
| 23 | RAD17 | 1551 | 2.152 | 0.3703 | Yes | ||
| 24 | CHFR | 1658 | 2.095 | 0.3792 | Yes | ||
| 25 | ZW10 | 1798 | 2.028 | 0.3859 | Yes | ||
| 26 | RAD51 | 1890 | 1.978 | 0.3949 | Yes | ||
| 27 | MPHOSPH6 | 2160 | 1.833 | 0.3932 | Yes | ||
| 28 | CDC25C | 2318 | 1.762 | 0.3970 | Yes | ||
| 29 | ZWINT | 2444 | 1.700 | 0.4022 | Yes | ||
| 30 | PIN1 | 2489 | 1.683 | 0.4116 | Yes | ||
| 31 | MRE11A | 2568 | 1.648 | 0.4189 | Yes | ||
| 32 | SMC4 | 2617 | 1.626 | 0.4277 | Yes | ||
| 33 | ANAPC5 | 2812 | 1.535 | 0.4280 | Yes | ||
| 34 | LIG3 | 2882 | 1.505 | 0.4348 | Yes | ||
| 35 | HSPA2 | 3319 | 1.325 | 0.4206 | Yes | ||
| 36 | SUGT1 | 3353 | 1.313 | 0.4280 | Yes | ||
| 37 | AURKA | 3558 | 1.229 | 0.4256 | Yes | ||
| 38 | PCBP4 | 3690 | 1.177 | 0.4267 | Yes | ||
| 39 | CIT | 3762 | 1.153 | 0.4310 | Yes | ||
| 40 | KNTC1 | 3770 | 1.150 | 0.4386 | Yes | ||
| 41 | CDC16 | 3837 | 1.128 | 0.4430 | Yes | ||
| 42 | EPGN | 4280 | 0.975 | 0.4259 | Yes | ||
| 43 | ANAPC10 | 4306 | 0.967 | 0.4314 | Yes | ||
| 44 | TTK | 4334 | 0.959 | 0.4366 | Yes | ||
| 45 | NPM2 | 4359 | 0.950 | 0.4420 | Yes | ||
| 46 | BIRC5 | 4482 | 0.913 | 0.4418 | Yes | ||
| 47 | ATM | 4489 | 0.911 | 0.4478 | Yes | ||
| 48 | CDC23 | 4716 | 0.834 | 0.4415 | No | ||
| 49 | DLG7 | 4982 | 0.762 | 0.4325 | No | ||
| 50 | RAN | 5137 | 0.712 | 0.4292 | No | ||
| 51 | XRCC2 | 5171 | 0.703 | 0.4323 | No | ||
| 52 | PPP5C | 5328 | 0.656 | 0.4285 | No | ||
| 53 | CHEK1 | 5824 | 0.517 | 0.4054 | No | ||
| 54 | TARDBP | 5888 | 0.503 | 0.4055 | No | ||
| 55 | KIF22 | 6386 | 0.378 | 0.3813 | No | ||
| 56 | NOLC1 | 6409 | 0.371 | 0.3827 | No | ||
| 57 | PAM | 6667 | 0.310 | 0.3710 | No | ||
| 58 | DDX11 | 6943 | 0.244 | 0.3579 | No | ||
| 59 | DCTN3 | 7240 | 0.178 | 0.3431 | No | ||
| 60 | KIF2C | 7420 | 0.138 | 0.3344 | No | ||
| 61 | CDCA5 | 7583 | 0.101 | 0.3264 | No | ||
| 62 | RAD54L | 7663 | 0.084 | 0.3227 | No | ||
| 63 | RAD52 | 7700 | 0.076 | 0.3213 | No | ||
| 64 | EREG | 7714 | 0.073 | 0.3211 | No | ||
| 65 | CCNA1 | 7930 | 0.025 | 0.3097 | No | ||
| 66 | UBE2C | 7954 | 0.017 | 0.3086 | No | ||
| 67 | BUB1 | 8427 | -0.087 | 0.2837 | No | ||
| 68 | RCC1 | 8994 | -0.206 | 0.2546 | No | ||
| 69 | PRMT5 | 9642 | -0.349 | 0.2221 | No | ||
| 70 | STAG3 | 9827 | -0.383 | 0.2148 | No | ||
| 71 | RAD1 | 9975 | -0.409 | 0.2098 | No | ||
| 72 | TGFB1 | 10258 | -0.463 | 0.1978 | No | ||
| 73 | MSH4 | 10342 | -0.479 | 0.1966 | No | ||
| 74 | CLIP1 | 10384 | -0.487 | 0.1978 | No | ||
| 75 | TOP3A | 11192 | -0.640 | 0.1587 | No | ||
| 76 | CD28 | 11391 | -0.675 | 0.1528 | No | ||
| 77 | KIF11 | 11471 | -0.690 | 0.1533 | No | ||
| 78 | NUMA1 | 11531 | -0.700 | 0.1551 | No | ||
| 79 | CDKN2B | 11760 | -0.733 | 0.1479 | No | ||
| 80 | DUSP13 | 11829 | -0.747 | 0.1495 | No | ||
| 81 | PIM2 | 12230 | -0.822 | 0.1336 | No | ||
| 82 | NEK6 | 12660 | -0.899 | 0.1167 | No | ||
| 83 | TTN | 13315 | -1.022 | 0.0886 | No | ||
| 84 | DMC1 | 14017 | -1.155 | 0.0588 | No | ||
| 85 | CETN1 | 14833 | -1.331 | 0.0241 | No | ||
| 86 | PLK1 | 15307 | -1.442 | 0.0087 | No | ||
| 87 | CENPE | 15495 | -1.491 | 0.0090 | No | ||
| 88 | CHMP1A | 15536 | -1.502 | 0.0174 | No | ||
| 89 | CDC25B | 15829 | -1.580 | 0.0127 | No | ||
| 90 | NEK2 | 15945 | -1.615 | 0.0178 | No | ||
| 91 | MAD2L2 | 16232 | -1.691 | 0.0142 | No | ||
| 92 | SSSCA1 | 16364 | -1.731 | 0.0192 | No | ||
| 93 | MSH5 | 16436 | -1.758 | 0.0277 | No | ||
| 94 | PML | 16972 | -1.966 | 0.0126 | No | ||
| 95 | DCTN2 | 17545 | -2.241 | -0.0026 | No | ||
| 96 | RAD51L3 | 17796 | -2.422 | 0.0009 | No | ||
| 97 | ANAPC11 | 17954 | -2.548 | 0.0102 | No | ||
| 98 | BOLL | 18517 | -3.635 | 0.0053 | No |
