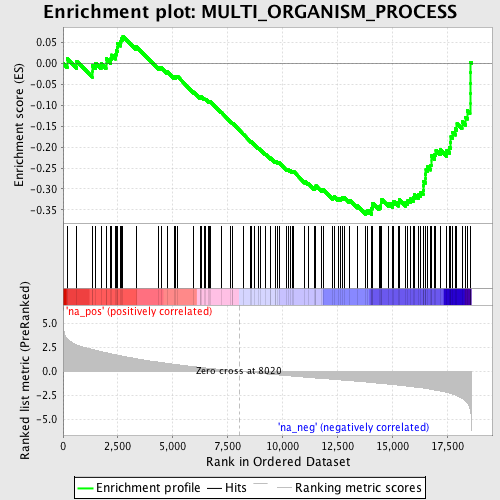
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
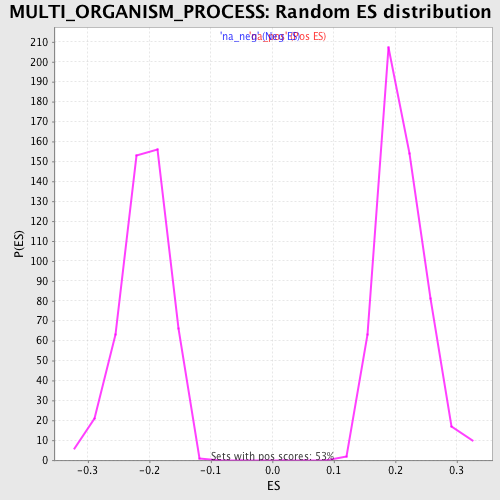
| GeneSet | MULTI_ORGANISM_PROCESS |
| Enrichment Score (ES) | -0.36088344 |
| Normalized Enrichment Score (NES) | -1.7378937 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.28258258 |
| FWER p-Value | 0.972 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CD4 | 184 | 3.407 | 0.0111 | No | ||
| 2 | OVGP1 | 616 | 2.765 | 0.0049 | No | ||
| 3 | FLT1 | 1323 | 2.287 | -0.0191 | No | ||
| 4 | SMC3 | 1328 | 2.281 | -0.0052 | No | ||
| 5 | PTPRC | 1485 | 2.196 | -0.0000 | No | ||
| 6 | IL12A | 1731 | 2.059 | -0.0005 | No | ||
| 7 | BCL3 | 1957 | 1.936 | -0.0007 | No | ||
| 8 | HNRPUL1 | 1959 | 1.935 | 0.0112 | No | ||
| 9 | NOD2 | 2179 | 1.826 | 0.0107 | No | ||
| 10 | ITCH | 2221 | 1.809 | 0.0196 | No | ||
| 11 | CHIA | 2402 | 1.721 | 0.0205 | No | ||
| 12 | IFNGR1 | 2422 | 1.713 | 0.0301 | No | ||
| 13 | IFNA7 | 2468 | 1.691 | 0.0381 | No | ||
| 14 | IVNS1ABP | 2484 | 1.685 | 0.0478 | No | ||
| 15 | TLR7 | 2611 | 1.629 | 0.0510 | No | ||
| 16 | SPRR2B | 2639 | 1.614 | 0.0595 | No | ||
| 17 | BNIP3L | 2726 | 1.573 | 0.0646 | No | ||
| 18 | TNF | 3329 | 1.321 | 0.0403 | No | ||
| 19 | TLR3 | 4362 | 0.950 | -0.0097 | No | ||
| 20 | CXCR4 | 4476 | 0.915 | -0.0101 | No | ||
| 21 | RRAGA | 4758 | 0.825 | -0.0202 | No | ||
| 22 | BCL10 | 5071 | 0.732 | -0.0325 | No | ||
| 23 | ADCYAP1 | 5136 | 0.712 | -0.0316 | No | ||
| 24 | INDO | 5201 | 0.694 | -0.0308 | No | ||
| 25 | MTNR1A | 5957 | 0.486 | -0.0686 | No | ||
| 26 | ACE2 | 6243 | 0.412 | -0.0814 | No | ||
| 27 | CRH | 6277 | 0.405 | -0.0807 | No | ||
| 28 | TLR8 | 6313 | 0.397 | -0.0801 | No | ||
| 29 | RSAD2 | 6450 | 0.363 | -0.0852 | No | ||
| 30 | NLRC4 | 6497 | 0.350 | -0.0856 | No | ||
| 31 | IFNAR2 | 6644 | 0.318 | -0.0915 | No | ||
| 32 | CD24 | 6690 | 0.305 | -0.0920 | No | ||
| 33 | CD81 | 6725 | 0.297 | -0.0920 | No | ||
| 34 | TEAD3 | 7219 | 0.184 | -0.1176 | No | ||
| 35 | CSH2 | 7645 | 0.088 | -0.1400 | No | ||
| 36 | CCL22 | 7712 | 0.073 | -0.1431 | No | ||
| 37 | SFRP4 | 7728 | 0.069 | -0.1435 | No | ||
| 38 | CRHBP | 8210 | -0.040 | -0.1692 | No | ||
| 39 | SPACA3 | 8562 | -0.118 | -0.1875 | No | ||
| 40 | HS3ST6 | 8582 | -0.122 | -0.1878 | No | ||
| 41 | PGLYRP3 | 8700 | -0.144 | -0.1932 | No | ||
| 42 | DUOX2 | 8896 | -0.185 | -0.2026 | No | ||
| 43 | CHIT1 | 9018 | -0.213 | -0.2078 | No | ||
| 44 | C9 | 9220 | -0.256 | -0.2171 | No | ||
| 45 | PZP | 9441 | -0.305 | -0.2271 | No | ||
| 46 | PRKRA | 9455 | -0.308 | -0.2259 | No | ||
| 47 | SOD1 | 9660 | -0.352 | -0.2348 | No | ||
| 48 | IFNAR1 | 9688 | -0.357 | -0.2340 | No | ||
| 49 | POLA1 | 9772 | -0.374 | -0.2362 | No | ||
| 50 | EPYC | 9842 | -0.385 | -0.2375 | No | ||
| 51 | FOSL1 | 10184 | -0.449 | -0.2532 | No | ||
| 52 | TGFB1 | 10258 | -0.463 | -0.2543 | No | ||
| 53 | BYSL | 10341 | -0.478 | -0.2557 | No | ||
| 54 | SPRR2G | 10450 | -0.497 | -0.2585 | No | ||
| 55 | DEFB103A | 10512 | -0.511 | -0.2586 | No | ||
| 56 | CREBZF | 11018 | -0.606 | -0.2822 | No | ||
| 57 | ADM | 11169 | -0.636 | -0.2864 | No | ||
| 58 | PTGFR | 11462 | -0.688 | -0.2979 | No | ||
| 59 | TRO | 11491 | -0.694 | -0.2951 | No | ||
| 60 | HBXIP | 11525 | -0.700 | -0.2926 | No | ||
| 61 | SCGB1A1 | 11786 | -0.737 | -0.3021 | No | ||
| 62 | SPRR2E | 11862 | -0.754 | -0.3015 | No | ||
| 63 | GHSR | 12294 | -0.834 | -0.3196 | No | ||
| 64 | SPRR2F | 12360 | -0.844 | -0.3179 | No | ||
| 65 | IL28RA | 12530 | -0.876 | -0.3216 | No | ||
| 66 | IFNGR2 | 12638 | -0.896 | -0.3218 | No | ||
| 67 | TARBP2 | 12716 | -0.910 | -0.3204 | No | ||
| 68 | FGR | 12807 | -0.927 | -0.3195 | No | ||
| 69 | CXCL12 | 13053 | -0.972 | -0.3267 | No | ||
| 70 | GHRL | 13403 | -1.039 | -0.3392 | No | ||
| 71 | PGLYRP1 | 13797 | -1.112 | -0.3535 | No | ||
| 72 | BNIP3 | 13886 | -1.129 | -0.3513 | No | ||
| 73 | CFP | 14064 | -1.165 | -0.3537 | Yes | ||
| 74 | IFNK | 14065 | -1.165 | -0.3465 | Yes | ||
| 75 | VAPB | 14092 | -1.171 | -0.3406 | Yes | ||
| 76 | CCL8 | 14100 | -1.174 | -0.3337 | Yes | ||
| 77 | LNPEP | 14400 | -1.243 | -0.3422 | Yes | ||
| 78 | COL16A1 | 14484 | -1.260 | -0.3389 | Yes | ||
| 79 | STAB1 | 14488 | -1.262 | -0.3313 | Yes | ||
| 80 | CCL5 | 14526 | -1.272 | -0.3254 | Yes | ||
| 81 | PPARD | 14841 | -1.333 | -0.3341 | Yes | ||
| 82 | SPRR2A | 15018 | -1.377 | -0.3351 | Yes | ||
| 83 | LALBA | 15066 | -1.387 | -0.3291 | Yes | ||
| 84 | RPL29 | 15284 | -1.436 | -0.3319 | Yes | ||
| 85 | CCL11 | 15314 | -1.443 | -0.3246 | Yes | ||
| 86 | PRLR | 15620 | -1.522 | -0.3316 | Yes | ||
| 87 | TLR6 | 15714 | -1.545 | -0.3271 | Yes | ||
| 88 | CAMP | 15832 | -1.582 | -0.3236 | Yes | ||
| 89 | IFNA4 | 15955 | -1.617 | -0.3202 | Yes | ||
| 90 | IRF7 | 16022 | -1.637 | -0.3137 | Yes | ||
| 91 | CRHR1 | 16211 | -1.685 | -0.3134 | Yes | ||
| 92 | SMAD3 | 16301 | -1.711 | -0.3076 | Yes | ||
| 93 | CLDN4 | 16408 | -1.748 | -0.3026 | Yes | ||
| 94 | FSHB | 16424 | -1.753 | -0.2925 | Yes | ||
| 95 | DMBT1 | 16433 | -1.756 | -0.2821 | Yes | ||
| 96 | CLIC5 | 16507 | -1.785 | -0.2750 | Yes | ||
| 97 | FCGRT | 16514 | -1.786 | -0.2643 | Yes | ||
| 98 | CCL4 | 16537 | -1.796 | -0.2543 | Yes | ||
| 99 | CCL19 | 16612 | -1.822 | -0.2471 | Yes | ||
| 100 | WFDC12 | 16757 | -1.878 | -0.2432 | Yes | ||
| 101 | LIPE | 16776 | -1.889 | -0.2325 | Yes | ||
| 102 | ISG20 | 16777 | -1.890 | -0.2208 | Yes | ||
| 103 | SPRR2D | 16942 | -1.954 | -0.2176 | Yes | ||
| 104 | HPGD | 16993 | -1.974 | -0.2081 | Yes | ||
| 105 | TNIP1 | 17192 | -2.054 | -0.2061 | Yes | ||
| 106 | PRF1 | 17489 | -2.210 | -0.2084 | Yes | ||
| 107 | PTHLH | 17605 | -2.275 | -0.2006 | Yes | ||
| 108 | DEFB127 | 17666 | -2.319 | -0.1895 | Yes | ||
| 109 | BANF1 | 17677 | -2.326 | -0.1756 | Yes | ||
| 110 | STAB2 | 17754 | -2.381 | -0.1650 | Yes | ||
| 111 | AGT | 17873 | -2.472 | -0.1561 | Yes | ||
| 112 | PGLYRP4 | 17951 | -2.545 | -0.1445 | Yes | ||
| 113 | SLC11A1 | 18182 | -2.808 | -0.1396 | Yes | ||
| 114 | ABCE1 | 18337 | -3.086 | -0.1288 | Yes | ||
| 115 | IL10 | 18421 | -3.274 | -0.1130 | Yes | ||
| 116 | BCL2 | 18546 | -3.877 | -0.0957 | Yes | ||
| 117 | PGLYRP2 | 18549 | -3.916 | -0.0716 | Yes | ||
| 118 | SPN | 18555 | -3.979 | -0.0473 | Yes | ||
| 119 | NOD1 | 18561 | -4.040 | -0.0225 | Yes | ||
| 120 | MAFF | 18569 | -4.115 | 0.0025 | Yes |