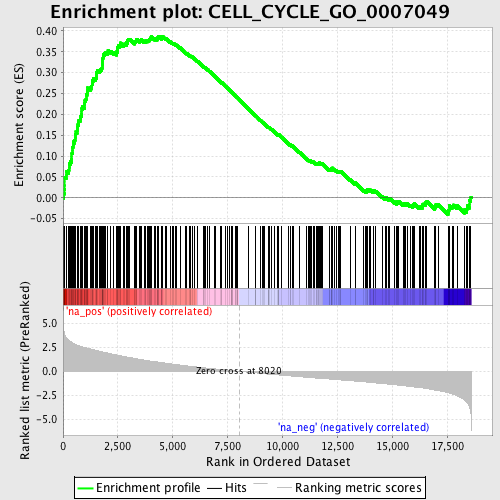
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CELL_CYCLE_GO_0007049 |
| Enrichment Score (ES) | 0.3874195 |
| Normalized Enrichment Score (NES) | 2.0453525 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008329735 |
| FWER p-Value | 0.123 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MADD | 21 | 4.378 | 0.0103 | Yes | ||
| 2 | TGFA | 49 | 4.065 | 0.0195 | Yes | ||
| 3 | PCAF | 66 | 3.924 | 0.0289 | Yes | ||
| 4 | ANLN | 70 | 3.873 | 0.0389 | Yes | ||
| 5 | CCND1 | 71 | 3.865 | 0.0490 | Yes | ||
| 6 | NDC80 | 138 | 3.521 | 0.0546 | Yes | ||
| 7 | LATS2 | 145 | 3.501 | 0.0635 | Yes | ||
| 8 | NCAPH | 260 | 3.228 | 0.0657 | Yes | ||
| 9 | APC | 270 | 3.202 | 0.0736 | Yes | ||
| 10 | SMC1A | 277 | 3.191 | 0.0817 | Yes | ||
| 11 | CUL4A | 332 | 3.111 | 0.0869 | Yes | ||
| 12 | CCNA2 | 386 | 3.023 | 0.0919 | Yes | ||
| 13 | HUS1 | 398 | 3.005 | 0.0992 | Yes | ||
| 14 | KIF15 | 401 | 2.995 | 0.1069 | Yes | ||
| 15 | RAD50 | 422 | 2.966 | 0.1136 | Yes | ||
| 16 | GTPBP4 | 427 | 2.963 | 0.1211 | Yes | ||
| 17 | CDC7 | 454 | 2.928 | 0.1274 | Yes | ||
| 18 | TPX2 | 474 | 2.908 | 0.1340 | Yes | ||
| 19 | RABGAP1 | 532 | 2.842 | 0.1383 | Yes | ||
| 20 | CHEK2 | 545 | 2.831 | 0.1451 | Yes | ||
| 21 | POLS | 554 | 2.824 | 0.1520 | Yes | ||
| 22 | EGF | 585 | 2.793 | 0.1577 | Yes | ||
| 23 | NUSAP1 | 636 | 2.749 | 0.1622 | Yes | ||
| 24 | SPO11 | 655 | 2.732 | 0.1684 | Yes | ||
| 25 | AKAP8 | 674 | 2.714 | 0.1745 | Yes | ||
| 26 | CUL3 | 707 | 2.691 | 0.1798 | Yes | ||
| 27 | MAD2L1 | 718 | 2.685 | 0.1863 | Yes | ||
| 28 | BUB1B | 774 | 2.637 | 0.1902 | Yes | ||
| 29 | SH3BP4 | 804 | 2.612 | 0.1955 | Yes | ||
| 30 | ATR | 839 | 2.586 | 0.2004 | Yes | ||
| 31 | STRN3 | 851 | 2.578 | 0.2066 | Yes | ||
| 32 | KPNA2 | 855 | 2.577 | 0.2131 | Yes | ||
| 33 | NASP | 872 | 2.569 | 0.2190 | Yes | ||
| 34 | GTF2H1 | 972 | 2.502 | 0.2202 | Yes | ||
| 35 | CCNT1 | 977 | 2.498 | 0.2265 | Yes | ||
| 36 | PRC1 | 994 | 2.488 | 0.2321 | Yes | ||
| 37 | POLE | 1038 | 2.455 | 0.2362 | Yes | ||
| 38 | HCFC1 | 1051 | 2.449 | 0.2420 | Yes | ||
| 39 | SYCP1 | 1056 | 2.444 | 0.2482 | Yes | ||
| 40 | ANAPC4 | 1093 | 2.423 | 0.2526 | Yes | ||
| 41 | KIF23 | 1105 | 2.417 | 0.2583 | Yes | ||
| 42 | CDK2 | 1128 | 2.401 | 0.2634 | Yes | ||
| 43 | BCCIP | 1238 | 2.335 | 0.2636 | Yes | ||
| 44 | RB1 | 1293 | 2.301 | 0.2666 | Yes | ||
| 45 | RAD51L1 | 1316 | 2.290 | 0.2714 | Yes | ||
| 46 | SMC3 | 1328 | 2.281 | 0.2768 | Yes | ||
| 47 | SESN1 | 1351 | 2.267 | 0.2816 | Yes | ||
| 48 | NBN | 1390 | 2.246 | 0.2854 | Yes | ||
| 49 | PTPRC | 1485 | 2.196 | 0.2860 | Yes | ||
| 50 | SKP2 | 1511 | 2.174 | 0.2903 | Yes | ||
| 51 | RAD21 | 1527 | 2.167 | 0.2952 | Yes | ||
| 52 | PAFAH1B1 | 1532 | 2.165 | 0.3006 | Yes | ||
| 53 | RAD17 | 1551 | 2.152 | 0.3053 | Yes | ||
| 54 | CHFR | 1658 | 2.095 | 0.3050 | Yes | ||
| 55 | NOTCH2 | 1715 | 2.067 | 0.3074 | Yes | ||
| 56 | CCNE2 | 1766 | 2.041 | 0.3100 | Yes | ||
| 57 | TBRG1 | 1777 | 2.036 | 0.3148 | Yes | ||
| 58 | CUL1 | 1780 | 2.035 | 0.3200 | Yes | ||
| 59 | EIF4G2 | 1793 | 2.030 | 0.3247 | Yes | ||
| 60 | ZW10 | 1798 | 2.028 | 0.3298 | Yes | ||
| 61 | CDKN2A | 1811 | 2.022 | 0.3344 | Yes | ||
| 62 | KHDRBS1 | 1822 | 2.018 | 0.3392 | Yes | ||
| 63 | UHRF2 | 1844 | 2.003 | 0.3433 | Yes | ||
| 64 | RAD51 | 1890 | 1.978 | 0.3460 | Yes | ||
| 65 | CETN3 | 1926 | 1.953 | 0.3492 | Yes | ||
| 66 | CDC45L | 2034 | 1.897 | 0.3483 | Yes | ||
| 67 | CCNK | 2044 | 1.893 | 0.3528 | Yes | ||
| 68 | MPHOSPH6 | 2160 | 1.833 | 0.3513 | Yes | ||
| 69 | CDC25C | 2318 | 1.762 | 0.3474 | Yes | ||
| 70 | FBXO5 | 2420 | 1.714 | 0.3464 | Yes | ||
| 71 | ZWINT | 2444 | 1.700 | 0.3496 | Yes | ||
| 72 | CCNT2 | 2466 | 1.693 | 0.3529 | Yes | ||
| 73 | CDKN1C | 2477 | 1.686 | 0.3568 | Yes | ||
| 74 | PIN1 | 2489 | 1.683 | 0.3606 | Yes | ||
| 75 | USP16 | 2502 | 1.679 | 0.3643 | Yes | ||
| 76 | MRE11A | 2568 | 1.648 | 0.3651 | Yes | ||
| 77 | GSPT1 | 2613 | 1.629 | 0.3670 | Yes | ||
| 78 | SMC4 | 2617 | 1.626 | 0.3710 | Yes | ||
| 79 | NDE1 | 2774 | 1.552 | 0.3666 | Yes | ||
| 80 | ANAPC5 | 2812 | 1.535 | 0.3686 | Yes | ||
| 81 | GMNN | 2881 | 1.506 | 0.3689 | Yes | ||
| 82 | LIG3 | 2882 | 1.505 | 0.3728 | Yes | ||
| 83 | MYH10 | 2933 | 1.483 | 0.3740 | Yes | ||
| 84 | CDT1 | 2942 | 1.479 | 0.3774 | Yes | ||
| 85 | PPP6C | 2973 | 1.463 | 0.3796 | Yes | ||
| 86 | USH1C | 3033 | 1.439 | 0.3802 | Yes | ||
| 87 | RACGAP1 | 3251 | 1.348 | 0.3719 | Yes | ||
| 88 | CCNG2 | 3257 | 1.345 | 0.3751 | Yes | ||
| 89 | HSPA2 | 3319 | 1.325 | 0.3753 | Yes | ||
| 90 | MNAT1 | 3322 | 1.324 | 0.3786 | Yes | ||
| 91 | SUGT1 | 3353 | 1.313 | 0.3804 | Yes | ||
| 92 | ERN1 | 3493 | 1.258 | 0.3761 | Yes | ||
| 93 | TIPIN | 3525 | 1.244 | 0.3777 | Yes | ||
| 94 | AURKA | 3558 | 1.229 | 0.3792 | Yes | ||
| 95 | PCBP4 | 3690 | 1.177 | 0.3751 | Yes | ||
| 96 | CIT | 3762 | 1.153 | 0.3743 | Yes | ||
| 97 | KNTC1 | 3770 | 1.150 | 0.3769 | Yes | ||
| 98 | CDC16 | 3837 | 1.128 | 0.3763 | Yes | ||
| 99 | DLG1 | 3892 | 1.106 | 0.3762 | Yes | ||
| 100 | CDK2AP1 | 3915 | 1.100 | 0.3779 | Yes | ||
| 101 | ERCC3 | 3940 | 1.088 | 0.3795 | Yes | ||
| 102 | GFI1B | 3964 | 1.077 | 0.3810 | Yes | ||
| 103 | CCNG1 | 3979 | 1.073 | 0.3831 | Yes | ||
| 104 | PTEN | 4007 | 1.063 | 0.3844 | Yes | ||
| 105 | UBB | 4015 | 1.059 | 0.3868 | Yes | ||
| 106 | ASNS | 4178 | 1.008 | 0.3806 | Yes | ||
| 107 | CDK5R1 | 4222 | 0.995 | 0.3809 | Yes | ||
| 108 | EPGN | 4280 | 0.975 | 0.3803 | Yes | ||
| 109 | ANAPC10 | 4306 | 0.967 | 0.3815 | Yes | ||
| 110 | CDC37 | 4318 | 0.963 | 0.3834 | Yes | ||
| 111 | TTK | 4334 | 0.959 | 0.3851 | Yes | ||
| 112 | NPM2 | 4359 | 0.950 | 0.3863 | Yes | ||
| 113 | GAS1 | 4480 | 0.913 | 0.3821 | Yes | ||
| 114 | BIRC5 | 4482 | 0.913 | 0.3845 | Yes | ||
| 115 | ATM | 4489 | 0.911 | 0.3865 | Yes | ||
| 116 | MYC | 4517 | 0.902 | 0.3874 | Yes | ||
| 117 | CDC6 | 4685 | 0.843 | 0.3805 | No | ||
| 118 | CDC23 | 4716 | 0.834 | 0.3811 | No | ||
| 119 | STMN1 | 4884 | 0.793 | 0.3741 | No | ||
| 120 | DLG7 | 4982 | 0.762 | 0.3708 | No | ||
| 121 | BRCA2 | 5040 | 0.743 | 0.3696 | No | ||
| 122 | RAN | 5137 | 0.712 | 0.3662 | No | ||
| 123 | XRCC2 | 5171 | 0.703 | 0.3663 | No | ||
| 124 | PPP5C | 5328 | 0.656 | 0.3595 | No | ||
| 125 | CDC25A | 5598 | 0.579 | 0.3464 | No | ||
| 126 | MFN2 | 5637 | 0.570 | 0.3458 | No | ||
| 127 | BMP4 | 5747 | 0.538 | 0.3413 | No | ||
| 128 | CDKN1A | 5813 | 0.522 | 0.3391 | No | ||
| 129 | CHEK1 | 5824 | 0.517 | 0.3399 | No | ||
| 130 | TARDBP | 5888 | 0.503 | 0.3378 | No | ||
| 131 | CCND3 | 6001 | 0.477 | 0.3329 | No | ||
| 132 | TUBE1 | 6134 | 0.443 | 0.3269 | No | ||
| 133 | KIF22 | 6386 | 0.378 | 0.3142 | No | ||
| 134 | NOLC1 | 6409 | 0.371 | 0.3140 | No | ||
| 135 | TUBG1 | 6454 | 0.363 | 0.3125 | No | ||
| 136 | GFI1 | 6478 | 0.356 | 0.3122 | No | ||
| 137 | XPC | 6597 | 0.329 | 0.3066 | No | ||
| 138 | PAM | 6667 | 0.310 | 0.3037 | No | ||
| 139 | MLF1 | 6887 | 0.259 | 0.2924 | No | ||
| 140 | DDX11 | 6943 | 0.244 | 0.2901 | No | ||
| 141 | MAP2K6 | 7150 | 0.203 | 0.2794 | No | ||
| 142 | MDM4 | 7239 | 0.178 | 0.2751 | No | ||
| 143 | DCTN3 | 7240 | 0.178 | 0.2755 | No | ||
| 144 | KIF2C | 7420 | 0.138 | 0.2661 | No | ||
| 145 | CENPF | 7488 | 0.120 | 0.2628 | No | ||
| 146 | CDCA5 | 7583 | 0.101 | 0.2579 | No | ||
| 147 | RAD54L | 7663 | 0.084 | 0.2538 | No | ||
| 148 | GPS2 | 7664 | 0.084 | 0.2541 | No | ||
| 149 | BTG3 | 7675 | 0.082 | 0.2537 | No | ||
| 150 | RAD52 | 7700 | 0.076 | 0.2526 | No | ||
| 151 | EREG | 7714 | 0.073 | 0.2521 | No | ||
| 152 | DHRS2 | 7837 | 0.047 | 0.2456 | No | ||
| 153 | GAS7 | 7872 | 0.037 | 0.2438 | No | ||
| 154 | JMY | 7917 | 0.026 | 0.2415 | No | ||
| 155 | CCNA1 | 7930 | 0.025 | 0.2409 | No | ||
| 156 | CCND2 | 7931 | 0.024 | 0.2410 | No | ||
| 157 | UBE2C | 7954 | 0.017 | 0.2398 | No | ||
| 158 | BUB1 | 8427 | -0.087 | 0.2143 | No | ||
| 159 | INHA | 8756 | -0.156 | 0.1969 | No | ||
| 160 | ALOX15B | 8776 | -0.159 | 0.1963 | No | ||
| 161 | CDKN2D | 8785 | -0.160 | 0.1962 | No | ||
| 162 | RCC1 | 8994 | -0.206 | 0.1854 | No | ||
| 163 | TP53 | 9109 | -0.232 | 0.1798 | No | ||
| 164 | PPP1R9B | 9138 | -0.238 | 0.1789 | No | ||
| 165 | PPM1G | 9178 | -0.246 | 0.1775 | No | ||
| 166 | RUNX3 | 9339 | -0.285 | 0.1695 | No | ||
| 167 | E2F1 | 9361 | -0.288 | 0.1691 | No | ||
| 168 | MCM2 | 9380 | -0.291 | 0.1689 | No | ||
| 169 | SERTAD1 | 9395 | -0.294 | 0.1689 | No | ||
| 170 | FANCG | 9504 | -0.321 | 0.1638 | No | ||
| 171 | PRMT5 | 9642 | -0.349 | 0.1573 | No | ||
| 172 | POLA1 | 9772 | -0.374 | 0.1512 | No | ||
| 173 | ANAPC2 | 9805 | -0.381 | 0.1505 | No | ||
| 174 | STAG3 | 9827 | -0.383 | 0.1504 | No | ||
| 175 | CKS2 | 9836 | -0.384 | 0.1509 | No | ||
| 176 | RAD1 | 9975 | -0.409 | 0.1445 | No | ||
| 177 | TGFB1 | 10258 | -0.463 | 0.1303 | No | ||
| 178 | MSH4 | 10342 | -0.479 | 0.1271 | No | ||
| 179 | CLIP1 | 10384 | -0.487 | 0.1261 | No | ||
| 180 | PA2G4 | 10433 | -0.495 | 0.1248 | No | ||
| 181 | NPM1 | 10521 | -0.513 | 0.1214 | No | ||
| 182 | SNF1LK | 10775 | -0.559 | 0.1091 | No | ||
| 183 | BMP2 | 11083 | -0.619 | 0.0940 | No | ||
| 184 | TOP3A | 11192 | -0.640 | 0.0897 | No | ||
| 185 | ACVR1 | 11232 | -0.648 | 0.0893 | No | ||
| 186 | FOXC1 | 11256 | -0.653 | 0.0898 | No | ||
| 187 | APBB2 | 11342 | -0.667 | 0.0869 | No | ||
| 188 | CD28 | 11391 | -0.675 | 0.0860 | No | ||
| 189 | KIF11 | 11471 | -0.690 | 0.0835 | No | ||
| 190 | NUMA1 | 11531 | -0.700 | 0.0822 | No | ||
| 191 | RHOB | 11550 | -0.704 | 0.0830 | No | ||
| 192 | HERC5 | 11587 | -0.709 | 0.0829 | No | ||
| 193 | CDK5RAP3 | 11646 | -0.719 | 0.0816 | No | ||
| 194 | CDKN3 | 11652 | -0.719 | 0.0833 | No | ||
| 195 | CENTD2 | 11675 | -0.722 | 0.0840 | No | ||
| 196 | EGFL6 | 11748 | -0.732 | 0.0819 | No | ||
| 197 | CDKN2B | 11760 | -0.733 | 0.0833 | No | ||
| 198 | DUSP13 | 11829 | -0.747 | 0.0815 | No | ||
| 199 | KATNA1 | 12149 | -0.807 | 0.0663 | No | ||
| 200 | CDK7 | 12156 | -0.809 | 0.0680 | No | ||
| 201 | TGFB2 | 12229 | -0.822 | 0.0663 | No | ||
| 202 | PIM2 | 12230 | -0.822 | 0.0684 | No | ||
| 203 | CDK5R2 | 12242 | -0.825 | 0.0700 | No | ||
| 204 | CITED2 | 12257 | -0.828 | 0.0714 | No | ||
| 205 | POLD1 | 12355 | -0.843 | 0.0683 | No | ||
| 206 | TIMELESS | 12455 | -0.862 | 0.0652 | No | ||
| 207 | STK11 | 12529 | -0.876 | 0.0635 | No | ||
| 208 | CDK6 | 12593 | -0.888 | 0.0624 | No | ||
| 209 | GADD45A | 12625 | -0.894 | 0.0630 | No | ||
| 210 | NEK6 | 12660 | -0.899 | 0.0635 | No | ||
| 211 | CUL5 | 13081 | -0.976 | 0.0432 | No | ||
| 212 | BTG4 | 13313 | -1.022 | 0.0333 | No | ||
| 213 | TTN | 13315 | -1.022 | 0.0359 | No | ||
| 214 | DDB1 | 13694 | -1.093 | 0.0182 | No | ||
| 215 | CDK5RAP1 | 13800 | -1.113 | 0.0154 | No | ||
| 216 | RPRM | 13845 | -1.121 | 0.0159 | No | ||
| 217 | TPD52L1 | 13858 | -1.124 | 0.0182 | No | ||
| 218 | RASSF1 | 13881 | -1.128 | 0.0200 | No | ||
| 219 | CUL2 | 13969 | -1.147 | 0.0182 | No | ||
| 220 | DMC1 | 14017 | -1.155 | 0.0187 | No | ||
| 221 | APBB1 | 14145 | -1.184 | 0.0149 | No | ||
| 222 | CDKN1B | 14151 | -1.185 | 0.0177 | No | ||
| 223 | MAP3K11 | 14248 | -1.207 | 0.0157 | No | ||
| 224 | ING4 | 14547 | -1.275 | 0.0028 | No | ||
| 225 | CDC20 | 14677 | -1.299 | -0.0009 | No | ||
| 226 | UHMK1 | 14719 | -1.305 | 0.0003 | No | ||
| 227 | CETN1 | 14833 | -1.331 | -0.0024 | No | ||
| 228 | SPHK1 | 14885 | -1.344 | -0.0016 | No | ||
| 229 | PFDN1 | 15102 | -1.395 | -0.0097 | No | ||
| 230 | PPP1R13B | 15216 | -1.419 | -0.0122 | No | ||
| 231 | AIF1 | 15237 | -1.424 | -0.0095 | No | ||
| 232 | PLK1 | 15307 | -1.442 | -0.0095 | No | ||
| 233 | CENPE | 15495 | -1.491 | -0.0158 | No | ||
| 234 | CHMP1A | 15536 | -1.502 | -0.0140 | No | ||
| 235 | GPS1 | 15604 | -1.517 | -0.0137 | No | ||
| 236 | ACVR1B | 15693 | -1.541 | -0.0145 | No | ||
| 237 | CDC25B | 15829 | -1.580 | -0.0177 | No | ||
| 238 | NEK2 | 15945 | -1.615 | -0.0197 | No | ||
| 239 | UPF1 | 15968 | -1.621 | -0.0167 | No | ||
| 240 | AHR | 16001 | -1.631 | -0.0141 | No | ||
| 241 | MAD2L2 | 16232 | -1.691 | -0.0223 | No | ||
| 242 | SMAD3 | 16301 | -1.711 | -0.0215 | No | ||
| 243 | SSSCA1 | 16364 | -1.731 | -0.0203 | No | ||
| 244 | MAPK12 | 16388 | -1.741 | -0.0170 | No | ||
| 245 | MSH5 | 16436 | -1.758 | -0.0150 | No | ||
| 246 | NEK11 | 16518 | -1.788 | -0.0147 | No | ||
| 247 | ZBTB17 | 16530 | -1.792 | -0.0106 | No | ||
| 248 | CDKN2C | 16578 | -1.811 | -0.0084 | No | ||
| 249 | RAD9A | 16939 | -1.953 | -0.0229 | No | ||
| 250 | PML | 16972 | -1.966 | -0.0195 | No | ||
| 251 | HPGD | 16993 | -1.974 | -0.0154 | No | ||
| 252 | BMP7 | 17093 | -2.014 | -0.0155 | No | ||
| 253 | DCTN2 | 17545 | -2.241 | -0.0343 | No | ||
| 254 | TBX3 | 17566 | -2.255 | -0.0294 | No | ||
| 255 | TBRG4 | 17590 | -2.266 | -0.0248 | No | ||
| 256 | DTYMK | 17592 | -2.267 | -0.0189 | No | ||
| 257 | ERCC2 | 17727 | -2.361 | -0.0200 | No | ||
| 258 | RAD51L3 | 17796 | -2.422 | -0.0173 | No | ||
| 259 | ANAPC11 | 17954 | -2.548 | -0.0192 | No | ||
| 260 | PLAGL1 | 18278 | -2.962 | -0.0291 | No | ||
| 261 | BCAT1 | 18395 | -3.210 | -0.0270 | No | ||
| 262 | ABL1 | 18411 | -3.245 | -0.0193 | No | ||
| 263 | CDK4 | 18513 | -3.623 | -0.0153 | No | ||
| 264 | BOLL | 18517 | -3.635 | -0.0059 | No | ||
| 265 | CDK10 | 18586 | -4.309 | 0.0016 | No |
