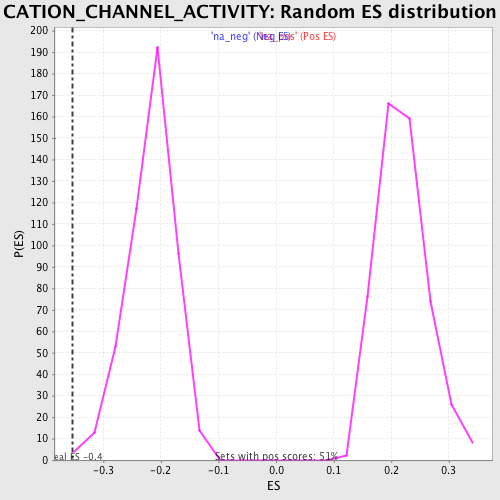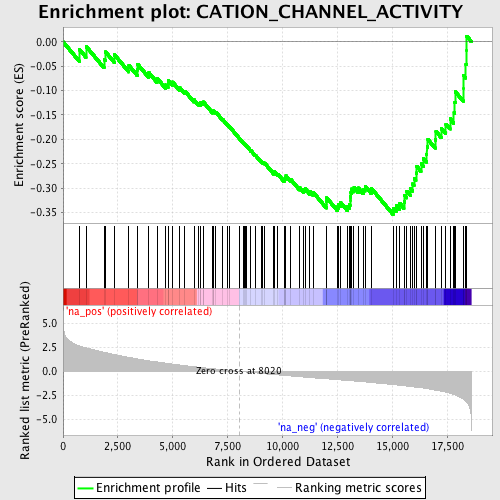
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CATION_CHANNEL_ACTIVITY |
| Enrichment Score (ES) | -0.3541512 |
| Normalized Enrichment Score (NES) | -1.6385561 |
| Nominal p-value | 0.0040899795 |
| FDR q-value | 0.40162355 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RYR3 | 747 | 2.664 | -0.0157 | No | ||
| 2 | SCN7A | 1045 | 2.451 | -0.0091 | No | ||
| 3 | HTR3B | 1869 | 1.988 | -0.0352 | No | ||
| 4 | CACNA1B | 1931 | 1.951 | -0.0205 | No | ||
| 5 | CHRNB2 | 2338 | 1.749 | -0.0262 | No | ||
| 6 | CACNG4 | 2998 | 1.453 | -0.0484 | No | ||
| 7 | CACNA1H | 3382 | 1.301 | -0.0571 | No | ||
| 8 | KCNH2 | 3399 | 1.293 | -0.0460 | No | ||
| 9 | RYR2 | 3909 | 1.101 | -0.0633 | No | ||
| 10 | KCNIP2 | 4282 | 0.974 | -0.0744 | No | ||
| 11 | CACNB1 | 4684 | 0.843 | -0.0882 | No | ||
| 12 | CHRNA4 | 4784 | 0.818 | -0.0860 | No | ||
| 13 | KCNJ3 | 4795 | 0.816 | -0.0790 | No | ||
| 14 | KCNA2 | 4986 | 0.762 | -0.0823 | No | ||
| 15 | NOLA1 | 5321 | 0.658 | -0.0942 | No | ||
| 16 | CHRNA1 | 5554 | 0.593 | -0.1013 | No | ||
| 17 | KCNJ15 | 5965 | 0.484 | -0.1189 | No | ||
| 18 | KCNMB2 | 6167 | 0.432 | -0.1258 | No | ||
| 19 | SCN5A | 6272 | 0.406 | -0.1277 | No | ||
| 20 | CACNB3 | 6273 | 0.405 | -0.1239 | No | ||
| 21 | PKD2 | 6376 | 0.380 | -0.1259 | No | ||
| 22 | KCNN4 | 6390 | 0.376 | -0.1231 | No | ||
| 23 | CACNB4 | 6817 | 0.275 | -0.1436 | No | ||
| 24 | CHRND | 6848 | 0.268 | -0.1427 | No | ||
| 25 | CACNA1A | 6869 | 0.263 | -0.1414 | No | ||
| 26 | KCNH3 | 6962 | 0.240 | -0.1441 | No | ||
| 27 | KCNK3 | 7267 | 0.171 | -0.1590 | No | ||
| 28 | KCNJ4 | 7501 | 0.116 | -0.1705 | No | ||
| 29 | CACNA1E | 7605 | 0.097 | -0.1751 | No | ||
| 30 | KCNK1 | 8017 | 0.001 | -0.1973 | No | ||
| 31 | KCNA1 | 8231 | -0.045 | -0.2084 | No | ||
| 32 | KCNA5 | 8285 | -0.055 | -0.2108 | No | ||
| 33 | KCNS3 | 8332 | -0.069 | -0.2126 | No | ||
| 34 | P2RX3 | 8340 | -0.071 | -0.2123 | No | ||
| 35 | KCNMB1 | 8561 | -0.118 | -0.2231 | No | ||
| 36 | SCN4A | 8765 | -0.157 | -0.2326 | No | ||
| 37 | KCNA4 | 9025 | -0.214 | -0.2446 | No | ||
| 38 | SCN11A | 9105 | -0.232 | -0.2468 | No | ||
| 39 | KCNJ5 | 9179 | -0.246 | -0.2484 | No | ||
| 40 | SCN9A | 9609 | -0.342 | -0.2684 | No | ||
| 41 | TRPC3 | 9615 | -0.343 | -0.2655 | No | ||
| 42 | KCNJ12 | 9785 | -0.376 | -0.2712 | No | ||
| 43 | KCNC1 | 10075 | -0.427 | -0.2828 | No | ||
| 44 | CACNG5 | 10107 | -0.434 | -0.2805 | No | ||
| 45 | KCND3 | 10133 | -0.439 | -0.2778 | No | ||
| 46 | RYR1 | 10156 | -0.443 | -0.2749 | No | ||
| 47 | KCNK7 | 10363 | -0.483 | -0.2816 | No | ||
| 48 | TRPC4 | 10779 | -0.560 | -0.2988 | No | ||
| 49 | MS4A2 | 10966 | -0.597 | -0.3033 | No | ||
| 50 | KCNMB4 | 11034 | -0.610 | -0.3013 | No | ||
| 51 | CHRNB4 | 11251 | -0.652 | -0.3070 | No | ||
| 52 | KCNQ3 | 11392 | -0.676 | -0.3083 | No | ||
| 53 | KCNC4 | 11998 | -0.781 | -0.3337 | No | ||
| 54 | TRPV6 | 12000 | -0.781 | -0.3266 | No | ||
| 55 | CACNA2D1 | 12012 | -0.783 | -0.3199 | No | ||
| 56 | CHRNE | 12505 | -0.871 | -0.3384 | No | ||
| 57 | CHRNA6 | 12560 | -0.882 | -0.3332 | No | ||
| 58 | CACNA1D | 12640 | -0.896 | -0.3292 | No | ||
| 59 | KCNJ10 | 12964 | -0.957 | -0.3378 | No | ||
| 60 | CACNA1G | 13054 | -0.972 | -0.3336 | No | ||
| 61 | CUL5 | 13081 | -0.976 | -0.3260 | No | ||
| 62 | TRPC1 | 13086 | -0.977 | -0.3172 | No | ||
| 63 | CHRNB3 | 13115 | -0.982 | -0.3096 | No | ||
| 64 | CHRNA2 | 13139 | -0.987 | -0.3018 | No | ||
| 65 | CACNG1 | 13256 | -1.011 | -0.2987 | No | ||
| 66 | KCNJ1 | 13454 | -1.048 | -0.2996 | No | ||
| 67 | KCNA3 | 13684 | -1.092 | -0.3019 | No | ||
| 68 | CACNA1S | 13760 | -1.105 | -0.2958 | No | ||
| 69 | KCNH1 | 14044 | -1.161 | -0.3003 | No | ||
| 70 | ACCN1 | 15042 | -1.381 | -0.3414 | Yes | ||
| 71 | P2RX1 | 15182 | -1.410 | -0.3359 | Yes | ||
| 72 | TRPM1 | 15344 | -1.450 | -0.3312 | Yes | ||
| 73 | SCNN1B | 15537 | -1.502 | -0.3277 | Yes | ||
| 74 | P2RX4 | 15566 | -1.508 | -0.3152 | Yes | ||
| 75 | TRPC5 | 15662 | -1.534 | -0.3062 | Yes | ||
| 76 | TRPM2 | 15836 | -1.583 | -0.3009 | Yes | ||
| 77 | SCNN1G | 15937 | -1.612 | -0.2914 | Yes | ||
| 78 | CHRNA7 | 15996 | -1.630 | -0.2795 | Yes | ||
| 79 | SCN1B | 16084 | -1.654 | -0.2689 | Yes | ||
| 80 | KCNE1 | 16120 | -1.662 | -0.2554 | Yes | ||
| 81 | CHRNA3 | 16313 | -1.715 | -0.2500 | Yes | ||
| 82 | KCNQ2 | 16422 | -1.752 | -0.2396 | Yes | ||
| 83 | KCNN2 | 16556 | -1.804 | -0.2301 | Yes | ||
| 84 | KCNC3 | 16585 | -1.813 | -0.2149 | Yes | ||
| 85 | KCNS1 | 16624 | -1.826 | -0.2001 | Yes | ||
| 86 | KCNQ1 | 16958 | -1.960 | -0.1999 | Yes | ||
| 87 | CACNA1C | 16992 | -1.973 | -0.1835 | Yes | ||
| 88 | CACNA1F | 17241 | -2.078 | -0.1777 | Yes | ||
| 89 | KCNK4 | 17446 | -2.180 | -0.1686 | Yes | ||
| 90 | KCNN3 | 17637 | -2.293 | -0.1576 | Yes | ||
| 91 | HTR3A | 17814 | -2.433 | -0.1447 | Yes | ||
| 92 | KCNE2 | 17853 | -2.462 | -0.1240 | Yes | ||
| 93 | KCNA6 | 17876 | -2.476 | -0.1023 | Yes | ||
| 94 | KCNJ14 | 18237 | -2.908 | -0.0949 | Yes | ||
| 95 | CACNB2 | 18241 | -2.911 | -0.0681 | Yes | ||
| 96 | KCNK5 | 18357 | -3.131 | -0.0454 | Yes | ||
| 97 | P2RX7 | 18392 | -3.205 | -0.0176 | Yes | ||
| 98 | KCNJ6 | 18397 | -3.212 | 0.0118 | Yes |