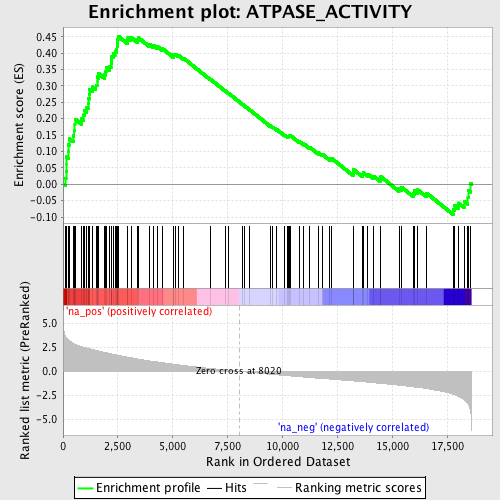
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ATPASE_ACTIVITY |
| Enrichment Score (ES) | 0.45209643 |
| Normalized Enrichment Score (NES) | 2.0748405 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0067958278 |
| FWER p-Value | 0.074 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDX20 | 97 | 3.704 | 0.0186 | Yes | ||
| 2 | ATP11B | 133 | 3.537 | 0.0395 | Yes | ||
| 3 | ATP2A2 | 151 | 3.491 | 0.0610 | Yes | ||
| 4 | CHD1 | 153 | 3.482 | 0.0834 | Yes | ||
| 5 | RECQL | 240 | 3.254 | 0.0997 | Yes | ||
| 6 | MPHOSPH1 | 246 | 3.248 | 0.1203 | Yes | ||
| 7 | DDX18 | 272 | 3.197 | 0.1396 | Yes | ||
| 8 | ATP7B | 460 | 2.924 | 0.1483 | Yes | ||
| 9 | SMARCA5 | 496 | 2.882 | 0.1650 | Yes | ||
| 10 | CLPX | 539 | 2.834 | 0.1809 | Yes | ||
| 11 | RSF1 | 542 | 2.833 | 0.1991 | Yes | ||
| 12 | G3BP1 | 841 | 2.585 | 0.1996 | Yes | ||
| 13 | DHX38 | 925 | 2.532 | 0.2114 | Yes | ||
| 14 | XRCC6 | 976 | 2.499 | 0.2248 | Yes | ||
| 15 | ATP1B3 | 1078 | 2.435 | 0.2351 | Yes | ||
| 16 | ATP6V1C1 | 1149 | 2.388 | 0.2466 | Yes | ||
| 17 | RALBP1 | 1165 | 2.381 | 0.2612 | Yes | ||
| 18 | ABCB7 | 1186 | 2.367 | 0.2753 | Yes | ||
| 19 | DDX21 | 1195 | 2.360 | 0.2901 | Yes | ||
| 20 | MYH9 | 1337 | 2.276 | 0.2971 | Yes | ||
| 21 | ABCD3 | 1498 | 2.185 | 0.3025 | Yes | ||
| 22 | DDX25 | 1548 | 2.153 | 0.3138 | Yes | ||
| 23 | VCP | 1550 | 2.152 | 0.3276 | Yes | ||
| 24 | CHD4 | 1626 | 2.116 | 0.3371 | Yes | ||
| 25 | RAD51 | 1890 | 1.978 | 0.3357 | Yes | ||
| 26 | ATP6V1B2 | 1940 | 1.944 | 0.3455 | Yes | ||
| 27 | TOP2A | 1966 | 1.933 | 0.3566 | Yes | ||
| 28 | ATP4A | 2128 | 1.849 | 0.3598 | Yes | ||
| 29 | SMARCAL1 | 2186 | 1.823 | 0.3685 | Yes | ||
| 30 | VPS4B | 2200 | 1.820 | 0.3795 | Yes | ||
| 31 | ATP1B1 | 2217 | 1.812 | 0.3903 | Yes | ||
| 32 | ATP1A1 | 2280 | 1.782 | 0.3984 | Yes | ||
| 33 | ABCF1 | 2366 | 1.733 | 0.4050 | Yes | ||
| 34 | ATP1B2 | 2430 | 1.708 | 0.4126 | Yes | ||
| 35 | ASCC3L1 | 2456 | 1.696 | 0.4222 | Yes | ||
| 36 | MSH6 | 2482 | 1.685 | 0.4317 | Yes | ||
| 37 | DDX3X | 2492 | 1.681 | 0.4420 | Yes | ||
| 38 | ABCC6 | 2506 | 1.678 | 0.4521 | Yes | ||
| 39 | MYH10 | 2933 | 1.483 | 0.4386 | No | ||
| 40 | FXYD2 | 2953 | 1.475 | 0.4471 | No | ||
| 41 | DHX9 | 3095 | 1.413 | 0.4486 | No | ||
| 42 | BPTF | 3388 | 1.299 | 0.4412 | No | ||
| 43 | PSMC6 | 3436 | 1.279 | 0.4469 | No | ||
| 44 | ERCC3 | 3940 | 1.088 | 0.4267 | No | ||
| 45 | PSMC5 | 4131 | 1.021 | 0.4231 | No | ||
| 46 | CCT8 | 4279 | 0.976 | 0.4214 | No | ||
| 47 | ATP2C1 | 4518 | 0.902 | 0.4144 | No | ||
| 48 | CHD2 | 5032 | 0.746 | 0.3915 | No | ||
| 49 | MYH7 | 5042 | 0.741 | 0.3958 | No | ||
| 50 | MYO9B | 5115 | 0.718 | 0.3965 | No | ||
| 51 | DDX56 | 5256 | 0.679 | 0.3933 | No | ||
| 52 | KIF1B | 5494 | 0.610 | 0.3844 | No | ||
| 53 | ATP7A | 6718 | 0.299 | 0.3203 | No | ||
| 54 | WRNIP1 | 7380 | 0.147 | 0.2856 | No | ||
| 55 | DHX8 | 7547 | 0.110 | 0.2773 | No | ||
| 56 | RFC3 | 8179 | -0.034 | 0.2435 | No | ||
| 57 | SMARCA1 | 8272 | -0.053 | 0.2389 | No | ||
| 58 | ABCA3 | 8506 | -0.106 | 0.2270 | No | ||
| 59 | MSH2 | 9439 | -0.304 | 0.1786 | No | ||
| 60 | ABCG2 | 9558 | -0.330 | 0.1743 | No | ||
| 61 | EIF4A3 | 9736 | -0.368 | 0.1672 | No | ||
| 62 | RUVBL1 | 10106 | -0.434 | 0.1500 | No | ||
| 63 | ATP6V0C | 10236 | -0.460 | 0.1460 | No | ||
| 64 | MYO1E | 10274 | -0.466 | 0.1470 | No | ||
| 65 | XRCC5 | 10318 | -0.473 | 0.1478 | No | ||
| 66 | MYO3A | 10357 | -0.481 | 0.1488 | No | ||
| 67 | ATP6V0E1 | 10778 | -0.560 | 0.1297 | No | ||
| 68 | ABCC1 | 10965 | -0.596 | 0.1235 | No | ||
| 69 | ABCC2 | 11228 | -0.648 | 0.1136 | No | ||
| 70 | ATP4B | 11630 | -0.716 | 0.0965 | No | ||
| 71 | PSMC1 | 11815 | -0.743 | 0.0914 | No | ||
| 72 | KATNA1 | 12149 | -0.807 | 0.0786 | No | ||
| 73 | ABCB11 | 12244 | -0.826 | 0.0788 | No | ||
| 74 | ATP2B3 | 13218 | -1.002 | 0.0327 | No | ||
| 75 | ABCD4 | 13237 | -1.007 | 0.0382 | No | ||
| 76 | PSMC4 | 13249 | -1.009 | 0.0441 | No | ||
| 77 | DDX54 | 13643 | -1.085 | 0.0299 | No | ||
| 78 | ABCG1 | 13670 | -1.089 | 0.0355 | No | ||
| 79 | ABCA2 | 13892 | -1.131 | 0.0309 | No | ||
| 80 | PPP2R4 | 14155 | -1.187 | 0.0244 | No | ||
| 81 | ATP1A2 | 14448 | -1.252 | 0.0167 | No | ||
| 82 | MYH6 | 14483 | -1.260 | 0.0229 | No | ||
| 83 | ABCC3 | 15311 | -1.443 | -0.0124 | No | ||
| 84 | RBBP4 | 15424 | -1.470 | -0.0090 | No | ||
| 85 | UPF1 | 15968 | -1.621 | -0.0279 | No | ||
| 86 | ATP2A1 | 16015 | -1.635 | -0.0198 | No | ||
| 87 | RUVBL2 | 16166 | -1.673 | -0.0172 | No | ||
| 88 | ATP1A3 | 16567 | -1.807 | -0.0271 | No | ||
| 89 | TAPBP | 17775 | -2.399 | -0.0769 | No | ||
| 90 | MYL6 | 17836 | -2.450 | -0.0643 | No | ||
| 91 | SKIV2L | 18018 | -2.619 | -0.0572 | No | ||
| 92 | PEX6 | 18295 | -2.993 | -0.0529 | No | ||
| 93 | PICK1 | 18452 | -3.360 | -0.0397 | No | ||
| 94 | ERCC8 | 18474 | -3.406 | -0.0189 | No | ||
| 95 | ATP2A3 | 18568 | -4.113 | 0.0026 | No |