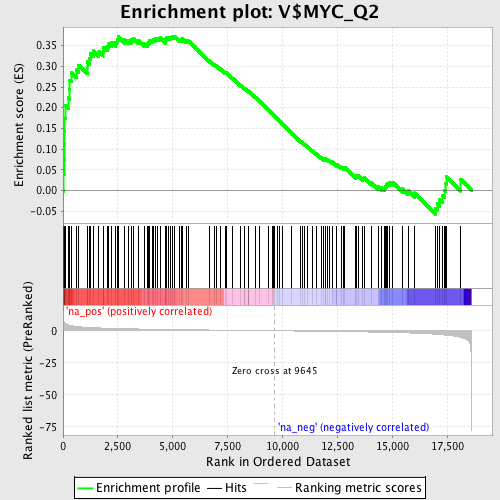
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.37306866 |
| Normalized Enrichment Score (NES) | 1.589131 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.088939734 |
| FWER p-Value | 0.624 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SHMT2 | 32 | 7.432 | 0.0389 | Yes | ||
| 2 | TIMM10 | 44 | 6.849 | 0.0757 | Yes | ||
| 3 | TAGLN2 | 57 | 6.453 | 0.1103 | Yes | ||
| 4 | HNRPA1 | 66 | 6.164 | 0.1436 | Yes | ||
| 5 | ATAD3A | 85 | 5.954 | 0.1751 | Yes | ||
| 6 | LDHA | 91 | 5.789 | 0.2065 | Yes | ||
| 7 | EBNA1BP2 | 236 | 4.558 | 0.2236 | Yes | ||
| 8 | PA2G4 | 296 | 4.312 | 0.2440 | Yes | ||
| 9 | CFL1 | 309 | 4.276 | 0.2667 | Yes | ||
| 10 | PFDN2 | 369 | 4.029 | 0.2856 | Yes | ||
| 11 | NUDC | 605 | 3.393 | 0.2914 | Yes | ||
| 12 | AMPD2 | 715 | 3.146 | 0.3027 | Yes | ||
| 13 | KRTCAP2 | 1101 | 2.642 | 0.2963 | Yes | ||
| 14 | PABPC4 | 1103 | 2.640 | 0.3107 | Yes | ||
| 15 | EIF4B | 1208 | 2.549 | 0.3190 | Yes | ||
| 16 | EIF3S10 | 1250 | 2.506 | 0.3305 | Yes | ||
| 17 | ADK | 1366 | 2.388 | 0.3373 | Yes | ||
| 18 | PPRC1 | 1634 | 2.205 | 0.3349 | Yes | ||
| 19 | AK2 | 1819 | 2.089 | 0.3364 | Yes | ||
| 20 | TIAL1 | 1850 | 2.070 | 0.3461 | Yes | ||
| 21 | B4GALT2 | 2010 | 1.980 | 0.3483 | Yes | ||
| 22 | HMGN2 | 2089 | 1.941 | 0.3547 | Yes | ||
| 23 | COPS7A | 2214 | 1.877 | 0.3583 | Yes | ||
| 24 | AP3M1 | 2400 | 1.797 | 0.3581 | Yes | ||
| 25 | RUSC1 | 2460 | 1.760 | 0.3645 | Yes | ||
| 26 | TFB2M | 2506 | 1.737 | 0.3716 | Yes | ||
| 27 | ATP5F1 | 2805 | 1.609 | 0.3642 | Yes | ||
| 28 | USP2 | 2979 | 1.544 | 0.3633 | Yes | ||
| 29 | RPL22 | 3113 | 1.500 | 0.3643 | Yes | ||
| 30 | CTBP2 | 3186 | 1.473 | 0.3685 | Yes | ||
| 31 | CDC14A | 3452 | 1.380 | 0.3617 | Yes | ||
| 32 | BCL9L | 3721 | 1.299 | 0.3543 | Yes | ||
| 33 | FKBP11 | 3852 | 1.258 | 0.3542 | Yes | ||
| 34 | PITX3 | 3898 | 1.240 | 0.3585 | Yes | ||
| 35 | GTF2H1 | 3951 | 1.227 | 0.3624 | Yes | ||
| 36 | IVNS1ABP | 4052 | 1.199 | 0.3635 | Yes | ||
| 37 | IL15RA | 4133 | 1.177 | 0.3657 | Yes | ||
| 38 | CNNM1 | 4216 | 1.154 | 0.3675 | Yes | ||
| 39 | FABP3 | 4316 | 1.132 | 0.3684 | Yes | ||
| 40 | SYT6 | 4415 | 1.106 | 0.3691 | Yes | ||
| 41 | SFXN2 | 4662 | 1.046 | 0.3615 | Yes | ||
| 42 | PLA2G4A | 4682 | 1.041 | 0.3662 | Yes | ||
| 43 | RAB3IL1 | 4732 | 1.029 | 0.3692 | Yes | ||
| 44 | SC5DL | 4805 | 1.013 | 0.3708 | Yes | ||
| 45 | TAF6L | 4911 | 0.989 | 0.3705 | Yes | ||
| 46 | PSEN2 | 4964 | 0.978 | 0.3731 | Yes | ||
| 47 | GPD1 | 5071 | 0.954 | 0.3726 | No | ||
| 48 | AGMAT | 5309 | 0.889 | 0.3646 | No | ||
| 49 | HOXC12 | 5400 | 0.864 | 0.3645 | No | ||
| 50 | RTN4RL2 | 5431 | 0.858 | 0.3675 | No | ||
| 51 | COL2A1 | 5608 | 0.819 | 0.3625 | No | ||
| 52 | TESK2 | 5709 | 0.797 | 0.3614 | No | ||
| 53 | POGK | 6661 | 0.597 | 0.3133 | No | ||
| 54 | SLC1A7 | 6888 | 0.551 | 0.3041 | No | ||
| 55 | RFX5 | 6985 | 0.533 | 0.3018 | No | ||
| 56 | STMN1 | 7185 | 0.491 | 0.2937 | No | ||
| 57 | CGN | 7397 | 0.445 | 0.2847 | No | ||
| 58 | UBXD3 | 7440 | 0.437 | 0.2849 | No | ||
| 59 | NR0B2 | 7731 | 0.382 | 0.2713 | No | ||
| 60 | BHLHB3 | 8100 | 0.303 | 0.2530 | No | ||
| 61 | LIN28 | 8250 | 0.271 | 0.2465 | No | ||
| 62 | LTBR | 8282 | 0.264 | 0.2462 | No | ||
| 63 | HNRPF | 8445 | 0.236 | 0.2388 | No | ||
| 64 | HOXC13 | 8448 | 0.236 | 0.2399 | No | ||
| 65 | B3GALT2 | 8462 | 0.232 | 0.2405 | No | ||
| 66 | RHEBL1 | 8774 | 0.169 | 0.2246 | No | ||
| 67 | CHRM1 | 8940 | 0.136 | 0.2165 | No | ||
| 68 | NRAS | 9380 | 0.052 | 0.1930 | No | ||
| 69 | B3GALT6 | 9557 | 0.016 | 0.1836 | No | ||
| 70 | SIRT1 | 9607 | 0.007 | 0.1810 | No | ||
| 71 | COMMD3 | 9620 | 0.005 | 0.1803 | No | ||
| 72 | CTSF | 9756 | -0.020 | 0.1732 | No | ||
| 73 | SLC16A1 | 9873 | -0.044 | 0.1671 | No | ||
| 74 | IPO13 | 9999 | -0.071 | 0.1608 | No | ||
| 75 | OPRD1 | 10414 | -0.150 | 0.1392 | No | ||
| 76 | NRIP3 | 10833 | -0.241 | 0.1179 | No | ||
| 77 | IPO7 | 10891 | -0.255 | 0.1162 | No | ||
| 78 | LHX9 | 11015 | -0.281 | 0.1111 | No | ||
| 79 | FADS3 | 11160 | -0.311 | 0.1050 | No | ||
| 80 | ALX4 | 11368 | -0.352 | 0.0958 | No | ||
| 81 | SLC6A15 | 11536 | -0.391 | 0.0889 | No | ||
| 82 | APOA5 | 11764 | -0.444 | 0.0790 | No | ||
| 83 | TDRD1 | 11878 | -0.468 | 0.0755 | No | ||
| 84 | HPCAL4 | 11943 | -0.478 | 0.0746 | No | ||
| 85 | WEE1 | 11945 | -0.478 | 0.0772 | No | ||
| 86 | NKX2-3 | 12049 | -0.500 | 0.0743 | No | ||
| 87 | USP15 | 12121 | -0.517 | 0.0733 | No | ||
| 88 | FOXD3 | 12260 | -0.547 | 0.0689 | No | ||
| 89 | HPCA | 12461 | -0.596 | 0.0613 | No | ||
| 90 | FOSL1 | 12479 | -0.600 | 0.0637 | No | ||
| 91 | TRIM46 | 12675 | -0.647 | 0.0567 | No | ||
| 92 | RRAGC | 12791 | -0.671 | 0.0541 | No | ||
| 93 | ATP6V0B | 12821 | -0.679 | 0.0563 | No | ||
| 94 | PAX6 | 13346 | -0.810 | 0.0324 | No | ||
| 95 | CHD4 | 13352 | -0.812 | 0.0365 | No | ||
| 96 | CENTB5 | 13455 | -0.837 | 0.0356 | No | ||
| 97 | POU3F1 | 13660 | -0.889 | 0.0294 | No | ||
| 98 | SCYL1 | 13721 | -0.904 | 0.0311 | No | ||
| 99 | H3F3A | 14056 | -1.000 | 0.0185 | No | ||
| 100 | ESRRA | 14352 | -1.089 | 0.0085 | No | ||
| 101 | ZZZ3 | 14501 | -1.135 | 0.0067 | No | ||
| 102 | PFKFB3 | 14652 | -1.183 | 0.0051 | No | ||
| 103 | CSDA | 14683 | -1.193 | 0.0100 | No | ||
| 104 | PTPRF | 14738 | -1.210 | 0.0137 | No | ||
| 105 | TGFB2 | 14788 | -1.230 | 0.0178 | No | ||
| 106 | ILK | 14888 | -1.271 | 0.0193 | No | ||
| 107 | LMX1A | 15006 | -1.319 | 0.0202 | No | ||
| 108 | LRP8 | 15469 | -1.537 | 0.0037 | No | ||
| 109 | ARL3 | 15722 | -1.676 | -0.0008 | No | ||
| 110 | FCHSD2 | 16009 | -1.849 | -0.0062 | No | ||
| 111 | ALDH3B1 | 16972 | -2.693 | -0.0435 | No | ||
| 112 | CBX5 | 17049 | -2.784 | -0.0324 | No | ||
| 113 | NIT1 | 17160 | -2.935 | -0.0223 | No | ||
| 114 | RORC | 17304 | -3.150 | -0.0128 | No | ||
| 115 | ATF7IP | 17385 | -3.293 | 0.0009 | No | ||
| 116 | FXYD2 | 17448 | -3.408 | 0.0162 | No | ||
| 117 | TMEM24 | 17465 | -3.435 | 0.0341 | No | ||
| 118 | FXYD6 | 18123 | -5.143 | 0.0266 | No |
