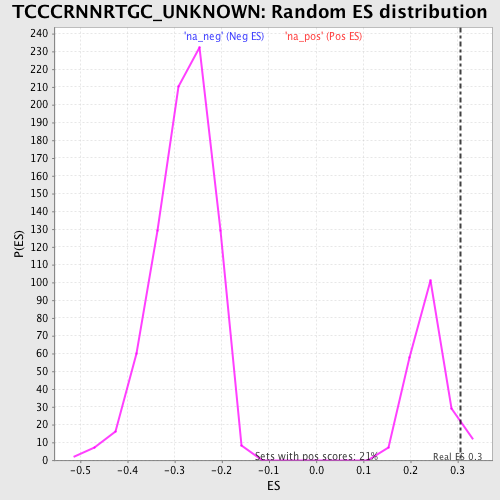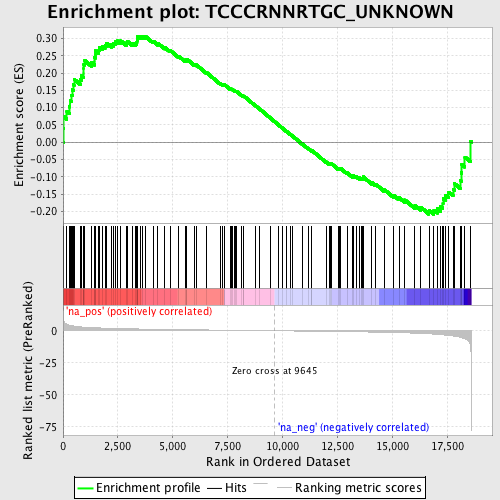
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TCCCRNNRTGC_UNKNOWN |
| Enrichment Score (ES) | 0.30681565 |
| Normalized Enrichment Score (NES) | 1.2886648 |
| Nominal p-value | 0.062801935 |
| FDR q-value | 0.4586823 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF5A | 17 | 8.994 | 0.0409 | Yes | ||
| 2 | ABCB6 | 35 | 7.229 | 0.0736 | Yes | ||
| 3 | POLR1A | 172 | 4.913 | 0.0891 | Yes | ||
| 4 | PA2G4 | 296 | 4.312 | 0.1025 | Yes | ||
| 5 | BUB1B | 313 | 4.263 | 0.1215 | Yes | ||
| 6 | GCN5L2 | 395 | 3.935 | 0.1354 | Yes | ||
| 7 | NUTF2 | 418 | 3.850 | 0.1522 | Yes | ||
| 8 | ORC4L | 476 | 3.719 | 0.1664 | Yes | ||
| 9 | IFRD2 | 501 | 3.645 | 0.1820 | Yes | ||
| 10 | RAD50 | 790 | 3.028 | 0.1806 | Yes | ||
| 11 | SFRS9 | 832 | 2.959 | 0.1921 | Yes | ||
| 12 | TCEB1 | 941 | 2.807 | 0.1993 | Yes | ||
| 13 | GART | 942 | 2.806 | 0.2124 | Yes | ||
| 14 | MIR16 | 946 | 2.803 | 0.2253 | Yes | ||
| 15 | HDAC8 | 989 | 2.756 | 0.2358 | Yes | ||
| 16 | SNX11 | 1294 | 2.454 | 0.2308 | Yes | ||
| 17 | NMT1 | 1440 | 2.327 | 0.2338 | Yes | ||
| 18 | SNX16 | 1441 | 2.327 | 0.2446 | Yes | ||
| 19 | RPL30 | 1466 | 2.307 | 0.2540 | Yes | ||
| 20 | USP16 | 1475 | 2.302 | 0.2643 | Yes | ||
| 21 | INSRR | 1628 | 2.208 | 0.2664 | Yes | ||
| 22 | MRPL50 | 1655 | 2.187 | 0.2751 | Yes | ||
| 23 | PPP1R8 | 1786 | 2.110 | 0.2779 | Yes | ||
| 24 | XPOT | 1912 | 2.036 | 0.2806 | Yes | ||
| 25 | VCL | 1975 | 2.000 | 0.2866 | Yes | ||
| 26 | SON | 2194 | 1.886 | 0.2836 | Yes | ||
| 27 | LIG1 | 2301 | 1.838 | 0.2864 | Yes | ||
| 28 | ACADVL | 2382 | 1.802 | 0.2905 | Yes | ||
| 29 | PSMD11 | 2485 | 1.748 | 0.2931 | Yes | ||
| 30 | MBP | 2632 | 1.678 | 0.2930 | Yes | ||
| 31 | RBM6 | 2877 | 1.586 | 0.2872 | Yes | ||
| 32 | EIF2B4 | 2921 | 1.568 | 0.2921 | Yes | ||
| 33 | VAPA | 3158 | 1.482 | 0.2863 | Yes | ||
| 34 | LZTR1 | 3294 | 1.436 | 0.2857 | Yes | ||
| 35 | SCAMP4 | 3359 | 1.409 | 0.2888 | Yes | ||
| 36 | POLK | 3368 | 1.406 | 0.2949 | Yes | ||
| 37 | UBADC1 | 3382 | 1.401 | 0.3007 | Yes | ||
| 38 | PCM1 | 3390 | 1.399 | 0.3068 | Yes | ||
| 39 | GSK3A | 3517 | 1.358 | 0.3063 | No | ||
| 40 | NOS3 | 3629 | 1.326 | 0.3065 | No | ||
| 41 | BRMS1 | 3738 | 1.295 | 0.3067 | No | ||
| 42 | MOSPD2 | 4101 | 1.186 | 0.2926 | No | ||
| 43 | KLK9 | 4321 | 1.130 | 0.2860 | No | ||
| 44 | CHCHD3 | 4628 | 1.054 | 0.2744 | No | ||
| 45 | RASSF6 | 4877 | 0.996 | 0.2656 | No | ||
| 46 | GRIN2B | 5276 | 0.898 | 0.2483 | No | ||
| 47 | PES1 | 5563 | 0.829 | 0.2367 | No | ||
| 48 | ZIC5 | 5611 | 0.818 | 0.2380 | No | ||
| 49 | TUBGCP3 | 5635 | 0.813 | 0.2405 | No | ||
| 50 | PLK1 | 6008 | 0.731 | 0.2238 | No | ||
| 51 | BSN | 6073 | 0.715 | 0.2237 | No | ||
| 52 | KPNB1 | 6534 | 0.622 | 0.2017 | No | ||
| 53 | STMN1 | 7185 | 0.491 | 0.1689 | No | ||
| 54 | RPP30 | 7259 | 0.474 | 0.1671 | No | ||
| 55 | TRIM23 | 7334 | 0.458 | 0.1652 | No | ||
| 56 | NEK2 | 7349 | 0.455 | 0.1666 | No | ||
| 57 | RRS1 | 7610 | 0.404 | 0.1544 | No | ||
| 58 | ASXL2 | 7654 | 0.396 | 0.1539 | No | ||
| 59 | PNRC2 | 7712 | 0.385 | 0.1527 | No | ||
| 60 | PCTK1 | 7833 | 0.360 | 0.1478 | No | ||
| 61 | TTC1 | 7872 | 0.351 | 0.1474 | No | ||
| 62 | SMPD3 | 7909 | 0.342 | 0.1471 | No | ||
| 63 | CKAP4 | 8133 | 0.296 | 0.1364 | No | ||
| 64 | RAB1A | 8219 | 0.278 | 0.1331 | No | ||
| 65 | MORF4L1 | 8233 | 0.275 | 0.1337 | No | ||
| 66 | TRAF3IP1 | 8759 | 0.172 | 0.1061 | No | ||
| 67 | POFUT1 | 8935 | 0.138 | 0.0973 | No | ||
| 68 | HSPH1 | 9463 | 0.035 | 0.0690 | No | ||
| 69 | RPL7 | 9805 | -0.028 | 0.0507 | No | ||
| 70 | RHOQ | 9991 | -0.069 | 0.0410 | No | ||
| 71 | NAP1L3 | 10172 | -0.104 | 0.0317 | No | ||
| 72 | VKORC1 | 10348 | -0.137 | 0.0229 | No | ||
| 73 | WTAP | 10433 | -0.154 | 0.0191 | No | ||
| 74 | IPO7 | 10891 | -0.255 | -0.0044 | No | ||
| 75 | SFRS1 | 11161 | -0.311 | -0.0175 | No | ||
| 76 | GSPT1 | 11321 | -0.345 | -0.0245 | No | ||
| 77 | EYA4 | 11326 | -0.346 | -0.0231 | No | ||
| 78 | BET1 | 11999 | -0.489 | -0.0572 | No | ||
| 79 | USP15 | 12121 | -0.517 | -0.0613 | No | ||
| 80 | NUDT13 | 12181 | -0.529 | -0.0620 | No | ||
| 81 | KIF11 | 12236 | -0.540 | -0.0624 | No | ||
| 82 | CSNK2A1 | 12552 | -0.617 | -0.0766 | No | ||
| 83 | COX4I1 | 12616 | -0.632 | -0.0770 | No | ||
| 84 | MAG | 12663 | -0.643 | -0.0765 | No | ||
| 85 | PHACTR3 | 12948 | -0.708 | -0.0886 | No | ||
| 86 | BRS3 | 13198 | -0.774 | -0.0985 | No | ||
| 87 | FUK | 13217 | -0.781 | -0.0958 | No | ||
| 88 | CHD4 | 13352 | -0.812 | -0.0993 | No | ||
| 89 | UBL3 | 13498 | -0.848 | -0.1032 | No | ||
| 90 | RB1CC1 | 13588 | -0.871 | -0.1039 | No | ||
| 91 | TIMP4 | 13661 | -0.890 | -0.1037 | No | ||
| 92 | DQX1 | 13671 | -0.892 | -0.1000 | No | ||
| 93 | SLC4A2 | 14068 | -1.003 | -0.1167 | No | ||
| 94 | RPS27A | 14252 | -1.060 | -0.1217 | No | ||
| 95 | SNX17 | 14654 | -1.184 | -0.1379 | No | ||
| 96 | TCTA | 15072 | -1.348 | -0.1541 | No | ||
| 97 | ISL1 | 15313 | -1.449 | -0.1604 | No | ||
| 98 | INVS | 15576 | -1.599 | -0.1671 | No | ||
| 99 | PUM1 | 16035 | -1.867 | -0.1832 | No | ||
| 100 | PRCC | 16299 | -2.064 | -0.1878 | No | ||
| 101 | IRF2BP1 | 16676 | -2.368 | -0.1971 | No | ||
| 102 | PMM2 | 16900 | -2.611 | -0.1970 | No | ||
| 103 | COL4A3BP | 17051 | -2.787 | -0.1921 | No | ||
| 104 | RDH10 | 17195 | -2.990 | -0.1859 | No | ||
| 105 | CNOT4 | 17313 | -3.166 | -0.1775 | No | ||
| 106 | ZCCHC8 | 17318 | -3.172 | -0.1630 | No | ||
| 107 | FYCO1 | 17437 | -3.383 | -0.1536 | No | ||
| 108 | CASC3 | 17576 | -3.645 | -0.1441 | No | ||
| 109 | CDK5 | 17776 | -4.065 | -0.1360 | No | ||
| 110 | PLAGL2 | 17833 | -4.192 | -0.1195 | No | ||
| 111 | FBS1 | 18099 | -5.050 | -0.1103 | No | ||
| 112 | MRPL24 | 18144 | -5.272 | -0.0882 | No | ||
| 113 | RHOA | 18172 | -5.441 | -0.0643 | No | ||
| 114 | NSD1 | 18314 | -6.234 | -0.0430 | No | ||
| 115 | TLE4 | 18570 | -12.734 | 0.0025 | No |