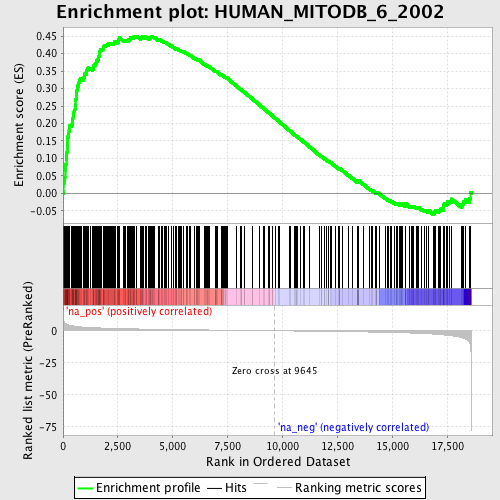
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HUMAN_MITODB_6_2002 |
| Enrichment Score (ES) | 0.45022258 |
| Normalized Enrichment Score (NES) | 2.2025483 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.8845237E-4 |
| FWER p-Value | 0.0040 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SHMT2 | 32 | 7.432 | 0.0088 | Yes | ||
| 2 | ABCB6 | 35 | 7.229 | 0.0189 | Yes | ||
| 3 | BAX | 38 | 6.993 | 0.0287 | Yes | ||
| 4 | TIMM10 | 44 | 6.849 | 0.0381 | Yes | ||
| 5 | MRPS10 | 59 | 6.411 | 0.0464 | Yes | ||
| 6 | ENDOG | 89 | 5.857 | 0.0531 | Yes | ||
| 7 | MRPS18B | 90 | 5.832 | 0.0614 | Yes | ||
| 8 | GOT2 | 92 | 5.775 | 0.0695 | Yes | ||
| 9 | SOD2 | 112 | 5.520 | 0.0762 | Yes | ||
| 10 | TIMM13 | 118 | 5.483 | 0.0837 | Yes | ||
| 11 | COX5A | 137 | 5.239 | 0.0902 | Yes | ||
| 12 | MRPL11 | 138 | 5.236 | 0.0976 | Yes | ||
| 13 | DHODH | 150 | 5.137 | 0.1042 | Yes | ||
| 14 | TXNRD2 | 162 | 5.034 | 0.1108 | Yes | ||
| 15 | CYC1 | 173 | 4.902 | 0.1171 | Yes | ||
| 16 | IMMT | 182 | 4.852 | 0.1236 | Yes | ||
| 17 | ACAA2 | 188 | 4.799 | 0.1301 | Yes | ||
| 18 | NDUFA10 | 194 | 4.757 | 0.1365 | Yes | ||
| 19 | ATP5G1 | 202 | 4.726 | 0.1429 | Yes | ||
| 20 | MRPS7 | 208 | 4.689 | 0.1492 | Yes | ||
| 21 | CLPP | 210 | 4.682 | 0.1558 | Yes | ||
| 22 | TOMM40 | 218 | 4.634 | 0.1620 | Yes | ||
| 23 | SLC25A3 | 234 | 4.571 | 0.1676 | Yes | ||
| 24 | MRPS15 | 259 | 4.452 | 0.1726 | Yes | ||
| 25 | MRPS30 | 267 | 4.434 | 0.1785 | Yes | ||
| 26 | ETFB | 281 | 4.358 | 0.1839 | Yes | ||
| 27 | MRPS28 | 288 | 4.334 | 0.1897 | Yes | ||
| 28 | IDH3A | 307 | 4.288 | 0.1948 | Yes | ||
| 29 | RPS3A | 386 | 3.975 | 0.1962 | Yes | ||
| 30 | HSPD1 | 415 | 3.873 | 0.2001 | Yes | ||
| 31 | MRPL12 | 419 | 3.849 | 0.2054 | Yes | ||
| 32 | PMPCB | 427 | 3.826 | 0.2104 | Yes | ||
| 33 | SHMT1 | 434 | 3.820 | 0.2155 | Yes | ||
| 34 | FPGS | 451 | 3.774 | 0.2200 | Yes | ||
| 35 | DLD | 461 | 3.746 | 0.2248 | Yes | ||
| 36 | ATP5G2 | 485 | 3.705 | 0.2288 | Yes | ||
| 37 | NDUFB8 | 495 | 3.672 | 0.2335 | Yes | ||
| 38 | ACADL | 505 | 3.635 | 0.2381 | Yes | ||
| 39 | ETFA | 540 | 3.558 | 0.2413 | Yes | ||
| 40 | SLC25A17 | 545 | 3.545 | 0.2461 | Yes | ||
| 41 | SUPV3L1 | 547 | 3.543 | 0.2511 | Yes | ||
| 42 | BCS1L | 563 | 3.501 | 0.2552 | Yes | ||
| 43 | MTCH2 | 567 | 3.494 | 0.2600 | Yes | ||
| 44 | UQCRC1 | 568 | 3.493 | 0.2649 | Yes | ||
| 45 | DNM1L | 578 | 3.463 | 0.2693 | Yes | ||
| 46 | ACADS | 593 | 3.424 | 0.2734 | Yes | ||
| 47 | SDHB | 594 | 3.417 | 0.2782 | Yes | ||
| 48 | MRPL4 | 596 | 3.416 | 0.2830 | Yes | ||
| 49 | MRPL13 | 604 | 3.399 | 0.2875 | Yes | ||
| 50 | ABCF2 | 615 | 3.369 | 0.2917 | Yes | ||
| 51 | HLCS | 629 | 3.326 | 0.2957 | Yes | ||
| 52 | MRPL19 | 647 | 3.287 | 0.2994 | Yes | ||
| 53 | MRPS27 | 663 | 3.251 | 0.3032 | Yes | ||
| 54 | MRPS18A | 668 | 3.244 | 0.3075 | Yes | ||
| 55 | MRPS11 | 695 | 3.185 | 0.3106 | Yes | ||
| 56 | ACAT1 | 696 | 3.182 | 0.3151 | Yes | ||
| 57 | COQ7 | 721 | 3.136 | 0.3182 | Yes | ||
| 58 | BID | 751 | 3.087 | 0.3210 | Yes | ||
| 59 | FDXR | 766 | 3.067 | 0.3246 | Yes | ||
| 60 | GOT1 | 780 | 3.042 | 0.3282 | Yes | ||
| 61 | TOMM22 | 836 | 2.951 | 0.3294 | Yes | ||
| 62 | SLC25A10 | 945 | 2.804 | 0.3274 | Yes | ||
| 63 | COX15 | 949 | 2.798 | 0.3312 | Yes | ||
| 64 | MRPS2 | 963 | 2.787 | 0.3344 | Yes | ||
| 65 | MRPS35 | 964 | 2.787 | 0.3384 | Yes | ||
| 66 | MTCH1 | 972 | 2.775 | 0.3419 | Yes | ||
| 67 | MDH2 | 1021 | 2.727 | 0.3432 | Yes | ||
| 68 | DBT | 1063 | 2.689 | 0.3447 | Yes | ||
| 69 | NDUFA8 | 1066 | 2.688 | 0.3484 | Yes | ||
| 70 | GSR | 1074 | 2.681 | 0.3518 | Yes | ||
| 71 | ATP5E | 1097 | 2.652 | 0.3544 | Yes | ||
| 72 | PCCB | 1121 | 2.623 | 0.3568 | Yes | ||
| 73 | NDUFB4 | 1134 | 2.608 | 0.3599 | Yes | ||
| 74 | RPL3 | 1229 | 2.528 | 0.3583 | Yes | ||
| 75 | ABCB7 | 1335 | 2.414 | 0.3560 | Yes | ||
| 76 | MRPL22 | 1378 | 2.378 | 0.3570 | Yes | ||
| 77 | SLC25A1 | 1379 | 2.378 | 0.3604 | Yes | ||
| 78 | TK1 | 1380 | 2.378 | 0.3638 | Yes | ||
| 79 | CYCS | 1401 | 2.360 | 0.3660 | Yes | ||
| 80 | MRPS18C | 1428 | 2.337 | 0.3679 | Yes | ||
| 81 | FIBP | 1471 | 2.304 | 0.3688 | Yes | ||
| 82 | OPA1 | 1488 | 2.293 | 0.3712 | Yes | ||
| 83 | MRPL48 | 1506 | 2.283 | 0.3735 | Yes | ||
| 84 | ATP5G3 | 1512 | 2.282 | 0.3765 | Yes | ||
| 85 | SURF1 | 1521 | 2.278 | 0.3793 | Yes | ||
| 86 | ATP7B | 1558 | 2.249 | 0.3805 | Yes | ||
| 87 | MRPS34 | 1582 | 2.232 | 0.3824 | Yes | ||
| 88 | PPID | 1593 | 2.224 | 0.3850 | Yes | ||
| 89 | SLC25A13 | 1606 | 2.217 | 0.3874 | Yes | ||
| 90 | CPT2 | 1615 | 2.212 | 0.3901 | Yes | ||
| 91 | VDAC2 | 1627 | 2.208 | 0.3927 | Yes | ||
| 92 | NDUFS7 | 1656 | 2.186 | 0.3942 | Yes | ||
| 93 | TXN2 | 1658 | 2.185 | 0.3973 | Yes | ||
| 94 | TRAP1 | 1659 | 2.184 | 0.4003 | Yes | ||
| 95 | TIMM17A | 1661 | 2.183 | 0.4034 | Yes | ||
| 96 | GLUD1 | 1676 | 2.172 | 0.4057 | Yes | ||
| 97 | MTRF1 | 1685 | 2.167 | 0.4083 | Yes | ||
| 98 | ATP5S | 1694 | 2.162 | 0.4109 | Yes | ||
| 99 | PYCR1 | 1725 | 2.144 | 0.4123 | Yes | ||
| 100 | TOMM70A | 1766 | 2.121 | 0.4131 | Yes | ||
| 101 | AK2 | 1819 | 2.089 | 0.4133 | Yes | ||
| 102 | PDHB | 1821 | 2.088 | 0.4162 | Yes | ||
| 103 | VDAC3 | 1826 | 2.080 | 0.4189 | Yes | ||
| 104 | NDUFC1 | 1858 | 2.064 | 0.4201 | Yes | ||
| 105 | UQCR | 1874 | 2.054 | 0.4222 | Yes | ||
| 106 | COX10 | 1906 | 2.037 | 0.4234 | Yes | ||
| 107 | HTATSF1 | 1947 | 2.012 | 0.4240 | Yes | ||
| 108 | MRPL34 | 1989 | 1.992 | 0.4246 | Yes | ||
| 109 | GCAT | 2025 | 1.970 | 0.4255 | Yes | ||
| 110 | PDK4 | 2076 | 1.949 | 0.4255 | Yes | ||
| 111 | COX6A2 | 2081 | 1.944 | 0.4280 | Yes | ||
| 112 | MRPL16 | 2124 | 1.927 | 0.4285 | Yes | ||
| 113 | ACAD8 | 2150 | 1.909 | 0.4298 | Yes | ||
| 114 | NDUFB5 | 2198 | 1.883 | 0.4299 | Yes | ||
| 115 | MRPL20 | 2267 | 1.853 | 0.4288 | Yes | ||
| 116 | ECHS1 | 2314 | 1.832 | 0.4289 | Yes | ||
| 117 | MAOA | 2325 | 1.826 | 0.4309 | Yes | ||
| 118 | MRPL46 | 2336 | 1.821 | 0.4329 | Yes | ||
| 119 | GPT | 2349 | 1.814 | 0.4348 | Yes | ||
| 120 | ACADVL | 2382 | 1.802 | 0.4356 | Yes | ||
| 121 | MRPS12 | 2483 | 1.749 | 0.4326 | Yes | ||
| 122 | ACP6 | 2490 | 1.745 | 0.4348 | Yes | ||
| 123 | ATP5D | 2522 | 1.729 | 0.4355 | Yes | ||
| 124 | AKAP1 | 2523 | 1.729 | 0.4380 | Yes | ||
| 125 | ACO2 | 2536 | 1.725 | 0.4397 | Yes | ||
| 126 | ALDH2 | 2547 | 1.719 | 0.4416 | Yes | ||
| 127 | MBD3 | 2565 | 1.710 | 0.4431 | Yes | ||
| 128 | TIMM23 | 2566 | 1.710 | 0.4455 | Yes | ||
| 129 | SLC25A4 | 2741 | 1.631 | 0.4383 | Yes | ||
| 130 | ATP5F1 | 2805 | 1.609 | 0.4372 | Yes | ||
| 131 | GPX4 | 2822 | 1.603 | 0.4385 | Yes | ||
| 132 | TUFM | 2864 | 1.589 | 0.4385 | Yes | ||
| 133 | NDUFAB1 | 2929 | 1.565 | 0.4373 | Yes | ||
| 134 | MTHFD1 | 2967 | 1.551 | 0.4374 | Yes | ||
| 135 | ATP5C1 | 2990 | 1.541 | 0.4384 | Yes | ||
| 136 | MFN1 | 2991 | 1.541 | 0.4406 | Yes | ||
| 137 | NME4 | 3033 | 1.530 | 0.4405 | Yes | ||
| 138 | OAT | 3053 | 1.521 | 0.4416 | Yes | ||
| 139 | UQCRB | 3055 | 1.521 | 0.4437 | Yes | ||
| 140 | BLVRA | 3061 | 1.519 | 0.4456 | Yes | ||
| 141 | TIMM8A | 3096 | 1.506 | 0.4459 | Yes | ||
| 142 | IDH3B | 3152 | 1.484 | 0.4450 | Yes | ||
| 143 | GATM | 3191 | 1.470 | 0.4450 | Yes | ||
| 144 | COX5B | 3201 | 1.468 | 0.4465 | Yes | ||
| 145 | SUCLA2 | 3217 | 1.464 | 0.4478 | Yes | ||
| 146 | NR3C1 | 3247 | 1.453 | 0.4483 | Yes | ||
| 147 | HSD3B2 | 3258 | 1.449 | 0.4498 | Yes | ||
| 148 | FECH | 3346 | 1.414 | 0.4470 | Yes | ||
| 149 | SDHD | 3348 | 1.413 | 0.4489 | Yes | ||
| 150 | CLN3 | 3362 | 1.408 | 0.4502 | Yes | ||
| 151 | SUCLG1 | 3530 | 1.354 | 0.4430 | No | ||
| 152 | ETFDH | 3545 | 1.350 | 0.4441 | No | ||
| 153 | MPST | 3557 | 1.346 | 0.4454 | No | ||
| 154 | HSPE1 | 3571 | 1.343 | 0.4466 | No | ||
| 155 | AMT | 3609 | 1.332 | 0.4465 | No | ||
| 156 | NDUFA6 | 3615 | 1.331 | 0.4481 | No | ||
| 157 | PDHX | 3621 | 1.330 | 0.4497 | No | ||
| 158 | MRPS31 | 3683 | 1.309 | 0.4482 | No | ||
| 159 | CNOT7 | 3775 | 1.284 | 0.4451 | No | ||
| 160 | NFS1 | 3789 | 1.281 | 0.4462 | No | ||
| 161 | NDUFS2 | 3794 | 1.278 | 0.4477 | No | ||
| 162 | SDHC | 3909 | 1.238 | 0.4433 | No | ||
| 163 | NDUFC2 | 3936 | 1.232 | 0.4436 | No | ||
| 164 | DLAT | 3947 | 1.228 | 0.4448 | No | ||
| 165 | NDUFB7 | 3966 | 1.223 | 0.4455 | No | ||
| 166 | UCP3 | 3995 | 1.216 | 0.4457 | No | ||
| 167 | PDHA1 | 4004 | 1.213 | 0.4470 | No | ||
| 168 | MRPS17 | 4011 | 1.212 | 0.4484 | No | ||
| 169 | IVNS1ABP | 4052 | 1.199 | 0.4479 | No | ||
| 170 | ATP5O | 4080 | 1.191 | 0.4481 | No | ||
| 171 | BAK1 | 4139 | 1.175 | 0.4466 | No | ||
| 172 | NDUFS8 | 4177 | 1.165 | 0.4462 | No | ||
| 173 | MRPS14 | 4326 | 1.129 | 0.4397 | No | ||
| 174 | ATP5J | 4340 | 1.126 | 0.4406 | No | ||
| 175 | MTERF | 4363 | 1.122 | 0.4409 | No | ||
| 176 | BAD | 4407 | 1.108 | 0.4402 | No | ||
| 177 | GALR3 | 4486 | 1.089 | 0.4374 | No | ||
| 178 | PIN4 | 4539 | 1.076 | 0.4361 | No | ||
| 179 | CYP27B1 | 4627 | 1.055 | 0.4328 | No | ||
| 180 | POPDC2 | 4667 | 1.045 | 0.4322 | No | ||
| 181 | HAX1 | 4723 | 1.032 | 0.4306 | No | ||
| 182 | MRPL33 | 4824 | 1.009 | 0.4266 | No | ||
| 183 | MFN2 | 4917 | 0.988 | 0.4229 | No | ||
| 184 | MRPL9 | 5041 | 0.960 | 0.4176 | No | ||
| 185 | COX7B | 5144 | 0.933 | 0.4133 | No | ||
| 186 | ALAS1 | 5145 | 0.933 | 0.4146 | No | ||
| 187 | HADHB | 5176 | 0.925 | 0.4143 | No | ||
| 188 | MRPL17 | 5244 | 0.907 | 0.4119 | No | ||
| 189 | GRID2 | 5308 | 0.890 | 0.4097 | No | ||
| 190 | MRPL35 | 5335 | 0.880 | 0.4095 | No | ||
| 191 | POLG2 | 5412 | 0.862 | 0.4066 | No | ||
| 192 | MAOB | 5417 | 0.860 | 0.4076 | No | ||
| 193 | ATP5I | 5475 | 0.848 | 0.4057 | No | ||
| 194 | CLPX | 5479 | 0.848 | 0.4067 | No | ||
| 195 | TAT | 5615 | 0.817 | 0.4005 | No | ||
| 196 | CYP27A1 | 5616 | 0.817 | 0.4016 | No | ||
| 197 | CS | 5656 | 0.810 | 0.4006 | No | ||
| 198 | PET112L | 5760 | 0.782 | 0.3961 | No | ||
| 199 | DECR1 | 5806 | 0.774 | 0.3947 | No | ||
| 200 | BCKDK | 5975 | 0.738 | 0.3866 | No | ||
| 201 | CPT1B | 5986 | 0.735 | 0.3871 | No | ||
| 202 | AGXT | 6062 | 0.717 | 0.3840 | No | ||
| 203 | HCLS1 | 6090 | 0.713 | 0.3835 | No | ||
| 204 | TIMM9 | 6130 | 0.705 | 0.3824 | No | ||
| 205 | SLC25A5 | 6145 | 0.703 | 0.3826 | No | ||
| 206 | SDS | 6170 | 0.697 | 0.3823 | No | ||
| 207 | PDK3 | 6193 | 0.691 | 0.3820 | No | ||
| 208 | OGG1 | 6448 | 0.637 | 0.3690 | No | ||
| 209 | DLST | 6498 | 0.629 | 0.3672 | No | ||
| 210 | COX6C | 6550 | 0.619 | 0.3653 | No | ||
| 211 | SLC1A3 | 6563 | 0.617 | 0.3655 | No | ||
| 212 | CYBA | 6577 | 0.614 | 0.3657 | No | ||
| 213 | SLC25A20 | 6643 | 0.600 | 0.3630 | No | ||
| 214 | HADHA | 6687 | 0.592 | 0.3615 | No | ||
| 215 | MTIF2 | 6935 | 0.543 | 0.3487 | No | ||
| 216 | PCCA | 6952 | 0.540 | 0.3486 | No | ||
| 217 | NDUFA5 | 6967 | 0.537 | 0.3486 | No | ||
| 218 | NDUFB2 | 6973 | 0.535 | 0.3491 | No | ||
| 219 | CRY1 | 6997 | 0.530 | 0.3486 | No | ||
| 220 | UQCRFS1 | 7023 | 0.524 | 0.3479 | No | ||
| 221 | BNIP3L | 7206 | 0.487 | 0.3387 | No | ||
| 222 | PRDX3 | 7212 | 0.484 | 0.3391 | No | ||
| 223 | MRPS16 | 7217 | 0.483 | 0.3395 | No | ||
| 224 | NDUFA9 | 7272 | 0.470 | 0.3373 | No | ||
| 225 | CKMT2 | 7331 | 0.458 | 0.3347 | No | ||
| 226 | SARDH | 7367 | 0.451 | 0.3335 | No | ||
| 227 | MRPS22 | 7418 | 0.441 | 0.3313 | No | ||
| 228 | MTHFD2 | 7423 | 0.440 | 0.3317 | No | ||
| 229 | MRPL39 | 7438 | 0.437 | 0.3316 | No | ||
| 230 | LYPLA2 | 7471 | 0.430 | 0.3305 | No | ||
| 231 | UQCRC2 | 7923 | 0.341 | 0.3062 | No | ||
| 232 | IDH3G | 8083 | 0.307 | 0.2980 | No | ||
| 233 | TIMM22 | 8126 | 0.298 | 0.2961 | No | ||
| 234 | HCCS | 8128 | 0.297 | 0.2965 | No | ||
| 235 | SLC25A28 | 8260 | 0.270 | 0.2897 | No | ||
| 236 | OTC | 8283 | 0.264 | 0.2888 | No | ||
| 237 | SLC25A15 | 8638 | 0.197 | 0.2697 | No | ||
| 238 | GCDH | 8652 | 0.194 | 0.2693 | No | ||
| 239 | ODC1 | 8937 | 0.137 | 0.2539 | No | ||
| 240 | NDUFS5 | 9118 | 0.105 | 0.2442 | No | ||
| 241 | CASP8 | 9176 | 0.093 | 0.2412 | No | ||
| 242 | MRPL18 | 9378 | 0.052 | 0.2303 | No | ||
| 243 | TOMM20 | 9406 | 0.048 | 0.2289 | No | ||
| 244 | CYP11B2 | 9425 | 0.043 | 0.2280 | No | ||
| 245 | HSD3B1 | 9529 | 0.020 | 0.2224 | No | ||
| 246 | NDUFA7 | 9541 | 0.018 | 0.2218 | No | ||
| 247 | IDH2 | 9687 | -0.009 | 0.2139 | No | ||
| 248 | PPIF | 9807 | -0.029 | 0.2074 | No | ||
| 249 | ATP5J2 | 9860 | -0.041 | 0.2046 | No | ||
| 250 | GLYAT | 10302 | -0.130 | 0.1806 | No | ||
| 251 | SDHA | 10362 | -0.140 | 0.1776 | No | ||
| 252 | HMGCS2 | 10557 | -0.180 | 0.1672 | No | ||
| 253 | HK1 | 10601 | -0.191 | 0.1652 | No | ||
| 254 | AFG3L2 | 10623 | -0.198 | 0.1643 | No | ||
| 255 | BCKDHB | 10656 | -0.205 | 0.1628 | No | ||
| 256 | COX7C | 10665 | -0.207 | 0.1627 | No | ||
| 257 | PPM2C | 10836 | -0.241 | 0.1537 | No | ||
| 258 | RPLP2 | 10960 | -0.267 | 0.1474 | No | ||
| 259 | ATPIF1 | 11000 | -0.279 | 0.1456 | No | ||
| 260 | BCKDHA | 11247 | -0.328 | 0.1326 | No | ||
| 261 | CPT1A | 11693 | -0.425 | 0.1088 | No | ||
| 262 | MRPL15 | 11756 | -0.442 | 0.1061 | No | ||
| 263 | CASQ1 | 11793 | -0.450 | 0.1047 | No | ||
| 264 | GCK | 11905 | -0.472 | 0.0993 | No | ||
| 265 | MRPL44 | 11935 | -0.476 | 0.0984 | No | ||
| 266 | MUT | 11987 | -0.484 | 0.0963 | No | ||
| 267 | COX11 | 12078 | -0.507 | 0.0921 | No | ||
| 268 | PPOX | 12093 | -0.511 | 0.0921 | No | ||
| 269 | MIPEP | 12117 | -0.516 | 0.0915 | No | ||
| 270 | ABAT | 12204 | -0.533 | 0.0876 | No | ||
| 271 | DNASE2 | 12215 | -0.537 | 0.0878 | No | ||
| 272 | ATP5H | 12426 | -0.587 | 0.0771 | No | ||
| 273 | ALDH4A1 | 12570 | -0.621 | 0.0702 | No | ||
| 274 | LARS2 | 12583 | -0.625 | 0.0704 | No | ||
| 275 | POLRMT | 12593 | -0.628 | 0.0708 | No | ||
| 276 | COX4I1 | 12616 | -0.632 | 0.0705 | No | ||
| 277 | GPD2 | 12712 | -0.655 | 0.0662 | No | ||
| 278 | BNIP3 | 12999 | -0.719 | 0.0516 | No | ||
| 279 | STARD3 | 13184 | -0.771 | 0.0426 | No | ||
| 280 | APAF1 | 13413 | -0.827 | 0.0313 | No | ||
| 281 | FDX1 | 13414 | -0.827 | 0.0324 | No | ||
| 282 | NDUFB3 | 13417 | -0.828 | 0.0335 | No | ||
| 283 | COX7A1 | 13427 | -0.829 | 0.0342 | No | ||
| 284 | SLC9A6 | 13447 | -0.835 | 0.0343 | No | ||
| 285 | FRAP1 | 13448 | -0.835 | 0.0355 | No | ||
| 286 | TIMM17B | 13468 | -0.840 | 0.0357 | No | ||
| 287 | TFB1M | 13472 | -0.842 | 0.0367 | No | ||
| 288 | NDUFA4 | 13684 | -0.895 | 0.0264 | No | ||
| 289 | TIMM44 | 13979 | -0.982 | 0.0117 | No | ||
| 290 | CRAT | 14063 | -1.001 | 0.0086 | No | ||
| 291 | TIMM8B | 14121 | -1.019 | 0.0069 | No | ||
| 292 | COX6A1 | 14255 | -1.060 | 0.0011 | No | ||
| 293 | ME2 | 14265 | -1.064 | 0.0021 | No | ||
| 294 | SLC25A12 | 14299 | -1.075 | 0.0018 | No | ||
| 295 | COX7A2 | 14304 | -1.077 | 0.0031 | No | ||
| 296 | MUTYH | 14426 | -1.111 | -0.0019 | No | ||
| 297 | ATP5B | 14686 | -1.194 | -0.0144 | No | ||
| 298 | TST | 14784 | -1.230 | -0.0180 | No | ||
| 299 | DAP3 | 14840 | -1.253 | -0.0192 | No | ||
| 300 | AMACR | 14906 | -1.277 | -0.0210 | No | ||
| 301 | MRPS33 | 14982 | -1.308 | -0.0232 | No | ||
| 302 | NDUFS3 | 15097 | -1.356 | -0.0276 | No | ||
| 303 | STAR | 15203 | -1.402 | -0.0313 | No | ||
| 304 | SPG7 | 15204 | -1.403 | -0.0293 | No | ||
| 305 | HSPA1L | 15247 | -1.421 | -0.0296 | No | ||
| 306 | NDUFA2 | 15344 | -1.467 | -0.0328 | No | ||
| 307 | OGDH | 15383 | -1.486 | -0.0328 | No | ||
| 308 | UQCRH | 15411 | -1.506 | -0.0321 | No | ||
| 309 | SLC25A16 | 15419 | -1.510 | -0.0304 | No | ||
| 310 | TOMM34 | 15458 | -1.530 | -0.0303 | No | ||
| 311 | CYP24A1 | 15588 | -1.603 | -0.0351 | No | ||
| 312 | ACADSB | 15609 | -1.613 | -0.0339 | No | ||
| 313 | ARG2 | 15612 | -1.614 | -0.0317 | No | ||
| 314 | NNT | 15616 | -1.616 | -0.0296 | No | ||
| 315 | PDK1 | 15809 | -1.722 | -0.0377 | No | ||
| 316 | HMGCL | 15879 | -1.761 | -0.0390 | No | ||
| 317 | UCP1 | 15908 | -1.778 | -0.0380 | No | ||
| 318 | RAF1 | 15947 | -1.805 | -0.0375 | No | ||
| 319 | MRP63 | 15990 | -1.839 | -0.0372 | No | ||
| 320 | NDUFS1 | 16100 | -1.909 | -0.0405 | No | ||
| 321 | SLC25A21 | 16152 | -1.955 | -0.0405 | No | ||
| 322 | CYBB | 16189 | -1.976 | -0.0397 | No | ||
| 323 | SLC1A1 | 16347 | -2.097 | -0.0453 | No | ||
| 324 | DGUOK | 16481 | -2.203 | -0.0495 | No | ||
| 325 | BCAT2 | 16540 | -2.245 | -0.0495 | No | ||
| 326 | SLC25A14 | 16641 | -2.336 | -0.0516 | No | ||
| 327 | TFAM | 16658 | -2.353 | -0.0492 | No | ||
| 328 | SCP2 | 16869 | -2.578 | -0.0570 | No | ||
| 329 | NDUFS4 | 16892 | -2.602 | -0.0545 | No | ||
| 330 | ACADM | 16942 | -2.663 | -0.0535 | No | ||
| 331 | NGFRAP1 | 16968 | -2.689 | -0.0510 | No | ||
| 332 | IVD | 16983 | -2.705 | -0.0480 | No | ||
| 333 | BCL2L11 | 17107 | -2.863 | -0.0506 | No | ||
| 334 | MRPL49 | 17154 | -2.925 | -0.0490 | No | ||
| 335 | POLG | 17186 | -2.973 | -0.0465 | No | ||
| 336 | UROS | 17222 | -3.039 | -0.0441 | No | ||
| 337 | MPO | 17324 | -3.184 | -0.0452 | No | ||
| 338 | CYP11A1 | 17333 | -3.202 | -0.0411 | No | ||
| 339 | COX7A2L | 17349 | -3.233 | -0.0373 | No | ||
| 340 | MCCC1 | 17353 | -3.236 | -0.0329 | No | ||
| 341 | TK2 | 17391 | -3.306 | -0.0302 | No | ||
| 342 | NDUFA3 | 17472 | -3.455 | -0.0297 | No | ||
| 343 | AKAP10 | 17508 | -3.523 | -0.0267 | No | ||
| 344 | BCL2L1 | 17522 | -3.558 | -0.0223 | No | ||
| 345 | DIABLO | 17625 | -3.729 | -0.0226 | No | ||
| 346 | NRF1 | 17703 | -3.905 | -0.0213 | No | ||
| 347 | PTPRA | 17712 | -3.927 | -0.0162 | No | ||
| 348 | MRPL24 | 18144 | -5.272 | -0.0324 | No | ||
| 349 | WARS2 | 18224 | -5.651 | -0.0287 | No | ||
| 350 | UCP2 | 18264 | -5.900 | -0.0225 | No | ||
| 351 | SLC25A11 | 18345 | -6.504 | -0.0176 | No | ||
| 352 | BCL2 | 18530 | -9.542 | -0.0142 | No | ||
| 353 | DCI | 18573 | -13.334 | 0.0024 | No |
