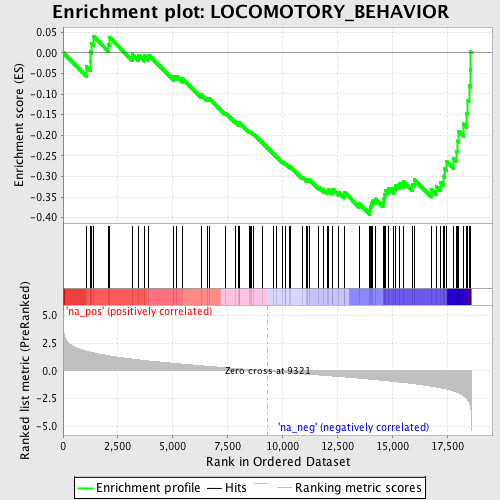
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | LOCOMOTORY_BEHAVIOR |
| Enrichment Score (ES) | -0.39229718 |
| Normalized Enrichment Score (NES) | -1.7368242 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.4094937 |
| FWER p-Value | 0.974 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ITGB2 | 1063 | 1.780 | -0.0334 | No | ||
| 2 | CXCR3 | 1242 | 1.697 | -0.0201 | No | ||
| 3 | SPN | 1247 | 1.695 | 0.0025 | No | ||
| 4 | FPR1 | 1308 | 1.669 | 0.0218 | No | ||
| 5 | HRAS | 1373 | 1.641 | 0.0404 | No | ||
| 6 | IL8RB | 2073 | 1.375 | 0.0212 | No | ||
| 7 | CCR6 | 2126 | 1.356 | 0.0367 | No | ||
| 8 | DEFB4 | 3149 | 1.071 | -0.0040 | No | ||
| 9 | CCL27 | 3429 | 1.007 | -0.0055 | No | ||
| 10 | CCL25 | 3699 | 0.945 | -0.0073 | No | ||
| 11 | CCL22 | 3908 | 0.899 | -0.0064 | No | ||
| 12 | RALA | 5026 | 0.691 | -0.0573 | No | ||
| 13 | CXCL1 | 5176 | 0.662 | -0.0564 | No | ||
| 14 | LTB4R2 | 5455 | 0.612 | -0.0632 | No | ||
| 15 | SOD1 | 6288 | 0.474 | -0.1017 | No | ||
| 16 | FGF2 | 6569 | 0.428 | -0.1110 | No | ||
| 17 | RNASE2 | 6669 | 0.413 | -0.1108 | No | ||
| 18 | IL4 | 7411 | 0.291 | -0.1469 | No | ||
| 19 | PROK2 | 7850 | 0.229 | -0.1674 | No | ||
| 20 | IL8RA | 8014 | 0.202 | -0.1735 | No | ||
| 21 | CCR3 | 8035 | 0.197 | -0.1719 | No | ||
| 22 | SCG2 | 8044 | 0.197 | -0.1697 | No | ||
| 23 | CCL19 | 8477 | 0.130 | -0.1912 | No | ||
| 24 | FOSL1 | 8533 | 0.121 | -0.1926 | No | ||
| 25 | CCR9 | 8580 | 0.112 | -0.1935 | No | ||
| 26 | CCL11 | 8680 | 0.096 | -0.1976 | No | ||
| 27 | CXCL10 | 9077 | 0.037 | -0.2184 | No | ||
| 28 | MAP2K1 | 9587 | -0.041 | -0.2453 | No | ||
| 29 | DEFB1 | 9704 | -0.063 | -0.2507 | No | ||
| 30 | XCL1 | 9997 | -0.114 | -0.2650 | No | ||
| 31 | SLIT2 | 10132 | -0.132 | -0.2704 | No | ||
| 32 | CXCL2 | 10145 | -0.134 | -0.2693 | No | ||
| 33 | SAA1 | 10339 | -0.162 | -0.2775 | No | ||
| 34 | CCL1 | 10358 | -0.166 | -0.2762 | No | ||
| 35 | DOCK2 | 10910 | -0.254 | -0.3025 | No | ||
| 36 | CXCL13 | 11111 | -0.285 | -0.3095 | No | ||
| 37 | NPY2R | 11152 | -0.291 | -0.3077 | No | ||
| 38 | CCR2 | 11221 | -0.302 | -0.3073 | No | ||
| 39 | C5AR1 | 11657 | -0.358 | -0.3260 | No | ||
| 40 | RALBP1 | 11881 | -0.393 | -0.3327 | No | ||
| 41 | MAPK14 | 12056 | -0.418 | -0.3364 | No | ||
| 42 | PLAUR | 12074 | -0.421 | -0.3317 | No | ||
| 43 | CXCL11 | 12280 | -0.450 | -0.3367 | No | ||
| 44 | CCR8 | 12297 | -0.452 | -0.3315 | No | ||
| 45 | LECT2 | 12571 | -0.494 | -0.3395 | No | ||
| 46 | MAPK1 | 12809 | -0.532 | -0.3451 | No | ||
| 47 | CCR7 | 12816 | -0.533 | -0.3383 | No | ||
| 48 | SCYE1 | 13493 | -0.647 | -0.3660 | No | ||
| 49 | CXCL12 | 13981 | -0.731 | -0.3824 | Yes | ||
| 50 | IL10 | 13995 | -0.733 | -0.3733 | Yes | ||
| 51 | CCL8 | 14044 | -0.742 | -0.3658 | Yes | ||
| 52 | CCRL1 | 14115 | -0.757 | -0.3594 | Yes | ||
| 53 | CCL24 | 14222 | -0.776 | -0.3547 | Yes | ||
| 54 | CCRL2 | 14581 | -0.845 | -0.3626 | Yes | ||
| 55 | FPRL1 | 14618 | -0.853 | -0.3531 | Yes | ||
| 56 | CCL2 | 14662 | -0.862 | -0.3438 | Yes | ||
| 57 | CCBP2 | 14703 | -0.870 | -0.3342 | Yes | ||
| 58 | PLD1 | 14833 | -0.899 | -0.3291 | Yes | ||
| 59 | SYK | 15062 | -0.946 | -0.3286 | Yes | ||
| 60 | CCL28 | 15163 | -0.964 | -0.3210 | Yes | ||
| 61 | PF4 | 15329 | -0.999 | -0.3165 | Yes | ||
| 62 | TGFB2 | 15506 | -1.035 | -0.3120 | Yes | ||
| 63 | CCL3 | 15932 | -1.138 | -0.3196 | Yes | ||
| 64 | CX3CL1 | 16001 | -1.156 | -0.3077 | Yes | ||
| 65 | CCL7 | 16786 | -1.368 | -0.3315 | Yes | ||
| 66 | XCR1 | 17014 | -1.442 | -0.3244 | Yes | ||
| 67 | CXCL5 | 17203 | -1.519 | -0.3140 | Yes | ||
| 68 | CXCL14 | 17354 | -1.585 | -0.3008 | Yes | ||
| 69 | CMKLR1 | 17393 | -1.601 | -0.2813 | Yes | ||
| 70 | SFTPD | 17473 | -1.631 | -0.2635 | Yes | ||
| 71 | CCR4 | 17789 | -1.799 | -0.2563 | Yes | ||
| 72 | CDH13 | 17941 | -1.902 | -0.2388 | Yes | ||
| 73 | CXCL9 | 17960 | -1.915 | -0.2140 | Yes | ||
| 74 | PIK3CB | 18011 | -1.964 | -0.1902 | Yes | ||
| 75 | CXCR4 | 18240 | -2.216 | -0.1727 | Yes | ||
| 76 | CX3CR1 | 18393 | -2.463 | -0.1477 | Yes | ||
| 77 | CCR5 | 18452 | -2.636 | -0.1153 | Yes | ||
| 78 | CCL5 | 18500 | -2.806 | -0.0800 | Yes | ||
| 79 | CCL17 | 18564 | -3.107 | -0.0416 | Yes | ||
| 80 | PLAU | 18579 | -3.288 | 0.0020 | Yes |
