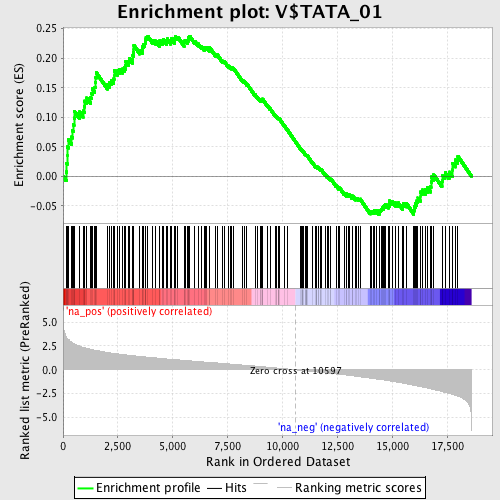
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$TATA_01 |
| Enrichment Score (ES) | 0.23746547 |
| Normalized Enrichment Score (NES) | 1.2226354 |
| Nominal p-value | 0.06185567 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KLF2 | 144 | 3.402 | 0.0076 | Yes | ||
| 2 | SCAMP3 | 166 | 3.350 | 0.0217 | Yes | ||
| 3 | HIST1H1A | 188 | 3.292 | 0.0355 | Yes | ||
| 4 | IL10 | 195 | 3.274 | 0.0500 | Yes | ||
| 5 | CSNK1E | 239 | 3.173 | 0.0621 | Yes | ||
| 6 | MAP2K5 | 395 | 2.871 | 0.0667 | Yes | ||
| 7 | PPARG | 433 | 2.809 | 0.0775 | Yes | ||
| 8 | PDYN | 476 | 2.755 | 0.0877 | Yes | ||
| 9 | HIST1H1B | 499 | 2.734 | 0.0989 | Yes | ||
| 10 | MRPS18B | 526 | 2.693 | 0.1097 | Yes | ||
| 11 | AGT | 743 | 2.472 | 0.1092 | Yes | ||
| 12 | PDLIM7 | 909 | 2.343 | 0.1109 | Yes | ||
| 13 | NPPA | 968 | 2.303 | 0.1182 | Yes | ||
| 14 | ASB4 | 988 | 2.289 | 0.1276 | Yes | ||
| 15 | MIP | 1081 | 2.237 | 0.1328 | Yes | ||
| 16 | ABLIM1 | 1252 | 2.137 | 0.1332 | Yes | ||
| 17 | GCH1 | 1294 | 2.113 | 0.1406 | Yes | ||
| 18 | EEF2 | 1329 | 2.099 | 0.1483 | Yes | ||
| 19 | POU3F4 | 1447 | 2.043 | 0.1512 | Yes | ||
| 20 | MMP10 | 1464 | 2.036 | 0.1596 | Yes | ||
| 21 | RPL10 | 1490 | 2.030 | 0.1674 | Yes | ||
| 22 | ADAMTS15 | 1510 | 2.023 | 0.1756 | Yes | ||
| 23 | CX3CL1 | 2026 | 1.816 | 0.1559 | Yes | ||
| 24 | TNFRSF17 | 2122 | 1.778 | 0.1588 | Yes | ||
| 25 | FSHB | 2192 | 1.753 | 0.1630 | Yes | ||
| 26 | NNT | 2301 | 1.716 | 0.1650 | Yes | ||
| 27 | BRUNOL4 | 2324 | 1.707 | 0.1715 | Yes | ||
| 28 | CLCN1 | 2330 | 1.705 | 0.1790 | Yes | ||
| 29 | HIST1H2BA | 2474 | 1.667 | 0.1788 | Yes | ||
| 30 | TCF7 | 2566 | 1.644 | 0.1813 | Yes | ||
| 31 | SMYD1 | 2691 | 1.607 | 0.1819 | Yes | ||
| 32 | CDC25B | 2787 | 1.580 | 0.1839 | Yes | ||
| 33 | COL10A1 | 2840 | 1.567 | 0.1882 | Yes | ||
| 34 | HNRPA0 | 2856 | 1.560 | 0.1945 | Yes | ||
| 35 | LIF | 2994 | 1.523 | 0.1939 | Yes | ||
| 36 | PCDH8 | 3017 | 1.517 | 0.1996 | Yes | ||
| 37 | TGM3 | 3167 | 1.477 | 0.1983 | Yes | ||
| 38 | CCL7 | 3176 | 1.474 | 0.2045 | Yes | ||
| 39 | UCN | 3204 | 1.467 | 0.2097 | Yes | ||
| 40 | HOXA10 | 3222 | 1.462 | 0.2154 | Yes | ||
| 41 | SOX5 | 3225 | 1.461 | 0.2219 | Yes | ||
| 42 | MYOZ2 | 3500 | 1.398 | 0.2134 | Yes | ||
| 43 | ZFP36L2 | 3606 | 1.375 | 0.2140 | Yes | ||
| 44 | NR4A1 | 3620 | 1.372 | 0.2195 | Yes | ||
| 45 | RBP2 | 3653 | 1.364 | 0.2239 | Yes | ||
| 46 | MYCT1 | 3732 | 1.344 | 0.2258 | Yes | ||
| 47 | TPM1 | 3743 | 1.341 | 0.2314 | Yes | ||
| 48 | COL1A1 | 3776 | 1.333 | 0.2357 | Yes | ||
| 49 | HOXC5 | 3861 | 1.315 | 0.2371 | Yes | ||
| 50 | CCL5 | 4090 | 1.272 | 0.2305 | Yes | ||
| 51 | CKM | 4214 | 1.244 | 0.2295 | Yes | ||
| 52 | PABPC4 | 4396 | 1.200 | 0.2251 | Yes | ||
| 53 | TSHB | 4410 | 1.197 | 0.2298 | Yes | ||
| 54 | SUPT4H1 | 4536 | 1.168 | 0.2283 | Yes | ||
| 55 | ECE2 | 4556 | 1.165 | 0.2326 | Yes | ||
| 56 | BNIP3 | 4730 | 1.129 | 0.2283 | Yes | ||
| 57 | SERPINH1 | 4739 | 1.127 | 0.2330 | Yes | ||
| 58 | HDLBP | 4909 | 1.096 | 0.2288 | Yes | ||
| 59 | CSH1 | 4920 | 1.093 | 0.2332 | Yes | ||
| 60 | TCTA | 5079 | 1.067 | 0.2295 | Yes | ||
| 61 | KRTAP21-1 | 5087 | 1.066 | 0.2340 | Yes | ||
| 62 | TNNC2 | 5120 | 1.056 | 0.2370 | Yes | ||
| 63 | HIST1H1T | 5230 | 1.036 | 0.2358 | Yes | ||
| 64 | DIRAS1 | 5514 | 0.980 | 0.2249 | Yes | ||
| 65 | NDRG2 | 5515 | 0.980 | 0.2293 | Yes | ||
| 66 | MT3 | 5578 | 0.968 | 0.2304 | Yes | ||
| 67 | SOX3 | 5686 | 0.950 | 0.2289 | Yes | ||
| 68 | KBTBD5 | 5699 | 0.947 | 0.2325 | Yes | ||
| 69 | ATOH1 | 5720 | 0.944 | 0.2357 | Yes | ||
| 70 | TTC17 | 5767 | 0.935 | 0.2375 | Yes | ||
| 71 | CRYBA1 | 6002 | 0.892 | 0.2288 | No | ||
| 72 | LHX6 | 6157 | 0.863 | 0.2244 | No | ||
| 73 | ABCA1 | 6317 | 0.834 | 0.2195 | No | ||
| 74 | FGFRL1 | 6430 | 0.813 | 0.2171 | No | ||
| 75 | S100A4 | 6474 | 0.805 | 0.2185 | No | ||
| 76 | AQP1 | 6532 | 0.797 | 0.2190 | No | ||
| 77 | RPS3 | 6667 | 0.770 | 0.2152 | No | ||
| 78 | RGS1 | 6677 | 0.769 | 0.2182 | No | ||
| 79 | HOXA11 | 6953 | 0.721 | 0.2066 | No | ||
| 80 | MT4 | 7017 | 0.712 | 0.2064 | No | ||
| 81 | BACH1 | 7275 | 0.667 | 0.1955 | No | ||
| 82 | LTA | 7341 | 0.657 | 0.1949 | No | ||
| 83 | CNN1 | 7538 | 0.619 | 0.1871 | No | ||
| 84 | DES | 7630 | 0.600 | 0.1849 | No | ||
| 85 | KRTAP6-2 | 7693 | 0.588 | 0.1842 | No | ||
| 86 | DKK2 | 7758 | 0.574 | 0.1833 | No | ||
| 87 | NPTX1 | 8175 | 0.496 | 0.1630 | No | ||
| 88 | RGS4 | 8249 | 0.483 | 0.1612 | No | ||
| 89 | CYP1A2 | 8376 | 0.461 | 0.1565 | No | ||
| 90 | GBE1 | 8745 | 0.390 | 0.1383 | No | ||
| 91 | CILP | 8857 | 0.372 | 0.1339 | No | ||
| 92 | ACTA1 | 9000 | 0.343 | 0.1278 | No | ||
| 93 | ADAM11 | 9011 | 0.341 | 0.1288 | No | ||
| 94 | TMEPAI | 9033 | 0.337 | 0.1292 | No | ||
| 95 | TRDN | 9065 | 0.329 | 0.1290 | No | ||
| 96 | SOSTDC1 | 9074 | 0.328 | 0.1300 | No | ||
| 97 | SLC34A1 | 9084 | 0.326 | 0.1310 | No | ||
| 98 | SPRR1B | 9332 | 0.274 | 0.1189 | No | ||
| 99 | ELAVL3 | 9432 | 0.248 | 0.1146 | No | ||
| 100 | HOXC4 | 9659 | 0.197 | 0.1032 | No | ||
| 101 | SCHIP1 | 9714 | 0.185 | 0.1011 | No | ||
| 102 | DUOX2 | 9720 | 0.185 | 0.1017 | No | ||
| 103 | OLIG2 | 9809 | 0.163 | 0.0977 | No | ||
| 104 | ESRRG | 9810 | 0.163 | 0.0984 | No | ||
| 105 | FGF16 | 9818 | 0.162 | 0.0988 | No | ||
| 106 | ADAMTS1 | 9877 | 0.151 | 0.0963 | No | ||
| 107 | TNFSF10 | 10079 | 0.113 | 0.0859 | No | ||
| 108 | PLUNC | 10218 | 0.082 | 0.0788 | No | ||
| 109 | CKB | 10816 | -0.055 | 0.0466 | No | ||
| 110 | GDF8 | 10847 | -0.061 | 0.0453 | No | ||
| 111 | DKK1 | 10890 | -0.069 | 0.0433 | No | ||
| 112 | MYH3 | 10921 | -0.078 | 0.0421 | No | ||
| 113 | BZW2 | 10925 | -0.078 | 0.0422 | No | ||
| 114 | CSH2 | 10971 | -0.088 | 0.0402 | No | ||
| 115 | HOXD4 | 11030 | -0.100 | 0.0375 | No | ||
| 116 | GADD45G | 11047 | -0.104 | 0.0371 | No | ||
| 117 | ARHGEF2 | 11096 | -0.114 | 0.0350 | No | ||
| 118 | WSB2 | 11107 | -0.115 | 0.0350 | No | ||
| 119 | SEPP1 | 11113 | -0.115 | 0.0353 | No | ||
| 120 | IL22 | 11130 | -0.120 | 0.0349 | No | ||
| 121 | TSC1 | 11354 | -0.172 | 0.0236 | No | ||
| 122 | CRABP2 | 11520 | -0.213 | 0.0156 | No | ||
| 123 | EGR2 | 11526 | -0.214 | 0.0163 | No | ||
| 124 | OLIG3 | 11530 | -0.215 | 0.0171 | No | ||
| 125 | BHLHB5 | 11553 | -0.218 | 0.0169 | No | ||
| 126 | SLC2A4 | 11623 | -0.234 | 0.0142 | No | ||
| 127 | MMP20 | 11625 | -0.234 | 0.0153 | No | ||
| 128 | PANK1 | 11734 | -0.261 | 0.0106 | No | ||
| 129 | ASB16 | 11746 | -0.263 | 0.0112 | No | ||
| 130 | CDX1 | 11754 | -0.264 | 0.0120 | No | ||
| 131 | NOG | 11937 | -0.308 | 0.0035 | No | ||
| 132 | NR0B2 | 12027 | -0.331 | 0.0002 | No | ||
| 133 | FGF20 | 12112 | -0.348 | -0.0028 | No | ||
| 134 | SCN8A | 12183 | -0.367 | -0.0049 | No | ||
| 135 | HIST1H1C | 12205 | -0.371 | -0.0044 | No | ||
| 136 | STAC | 12440 | -0.429 | -0.0151 | No | ||
| 137 | MYF6 | 12567 | -0.464 | -0.0199 | No | ||
| 138 | EDN1 | 12601 | -0.473 | -0.0195 | No | ||
| 139 | CA2 | 12818 | -0.525 | -0.0288 | No | ||
| 140 | DIO2 | 12913 | -0.549 | -0.0315 | No | ||
| 141 | IL11 | 12922 | -0.551 | -0.0294 | No | ||
| 142 | GRID2 | 13006 | -0.576 | -0.0313 | No | ||
| 143 | HIST1H2AA | 13056 | -0.592 | -0.0313 | No | ||
| 144 | FGF4 | 13178 | -0.623 | -0.0350 | No | ||
| 145 | RHOA | 13198 | -0.627 | -0.0332 | No | ||
| 146 | PLAG1 | 13330 | -0.670 | -0.0372 | No | ||
| 147 | MITF | 13381 | -0.686 | -0.0368 | No | ||
| 148 | PRDX1 | 13473 | -0.709 | -0.0386 | No | ||
| 149 | GSC | 13532 | -0.727 | -0.0384 | No | ||
| 150 | PURA | 14002 | -0.866 | -0.0599 | No | ||
| 151 | STC1 | 14067 | -0.889 | -0.0594 | No | ||
| 152 | CXCR4 | 14140 | -0.915 | -0.0591 | No | ||
| 153 | RLN3 | 14198 | -0.932 | -0.0580 | No | ||
| 154 | GPRC5D | 14281 | -0.959 | -0.0581 | No | ||
| 155 | JARID2 | 14403 | -0.998 | -0.0601 | No | ||
| 156 | RASGRP3 | 14432 | -1.005 | -0.0571 | No | ||
| 157 | EGFL7 | 14488 | -1.022 | -0.0554 | No | ||
| 158 | PTP4A1 | 14534 | -1.036 | -0.0532 | No | ||
| 159 | SLC25A4 | 14588 | -1.055 | -0.0513 | No | ||
| 160 | RUNX2 | 14630 | -1.071 | -0.0486 | No | ||
| 161 | GTF2A1 | 14698 | -1.099 | -0.0473 | No | ||
| 162 | NANOS1 | 14821 | -1.141 | -0.0487 | No | ||
| 163 | NEUROG2 | 14872 | -1.159 | -0.0462 | No | ||
| 164 | AMPD1 | 14885 | -1.163 | -0.0415 | No | ||
| 165 | FOXP3 | 14998 | -1.207 | -0.0421 | No | ||
| 166 | KIRREL2 | 15156 | -1.268 | -0.0449 | No | ||
| 167 | NEFH | 15266 | -1.313 | -0.0448 | No | ||
| 168 | RHOBTB1 | 15475 | -1.396 | -0.0498 | No | ||
| 169 | GRIN2B | 15514 | -1.411 | -0.0455 | No | ||
| 170 | TMPO | 15636 | -1.460 | -0.0454 | No | ||
| 171 | EEF1A1 | 15991 | -1.619 | -0.0573 | No | ||
| 172 | DDX5 | 16021 | -1.638 | -0.0514 | No | ||
| 173 | H2AFZ | 16050 | -1.649 | -0.0454 | No | ||
| 174 | DDX3X | 16124 | -1.681 | -0.0418 | No | ||
| 175 | MID1 | 16173 | -1.701 | -0.0366 | No | ||
| 176 | SIM1 | 16276 | -1.747 | -0.0342 | No | ||
| 177 | HOXD9 | 16279 | -1.749 | -0.0264 | No | ||
| 178 | CRYAB | 16367 | -1.793 | -0.0230 | No | ||
| 179 | BMI1 | 16510 | -1.860 | -0.0223 | No | ||
| 180 | HSPB2 | 16603 | -1.912 | -0.0186 | No | ||
| 181 | MAT2A | 16755 | -1.992 | -0.0177 | No | ||
| 182 | MYH4 | 16770 | -2.002 | -0.0094 | No | ||
| 183 | CFL2 | 16783 | -2.009 | -0.0009 | No | ||
| 184 | COL13A1 | 16892 | -2.063 | 0.0026 | No | ||
| 185 | ITPR1 | 17270 | -2.271 | -0.0076 | No | ||
| 186 | PPP1R10 | 17304 | -2.293 | 0.0010 | No | ||
| 187 | TTR | 17406 | -2.350 | 0.0062 | No | ||
| 188 | EIF4A2 | 17593 | -2.470 | 0.0073 | No | ||
| 189 | NASP | 17744 | -2.569 | 0.0109 | No | ||
| 190 | H3F3B | 17752 | -2.572 | 0.0222 | No | ||
| 191 | SFRS3 | 17871 | -2.665 | 0.0279 | No | ||
| 192 | ANKMY2 | 17993 | -2.757 | 0.0338 | No |
