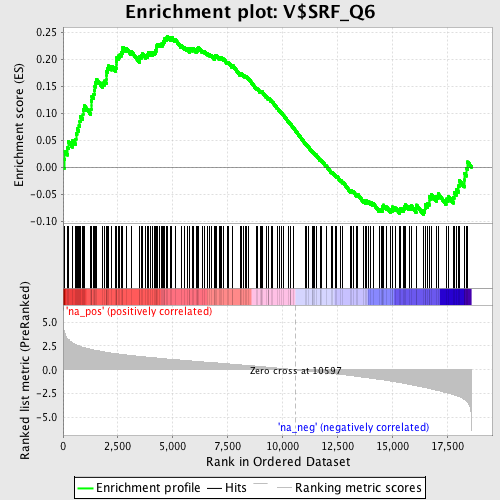
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$SRF_Q6 |
| Enrichment Score (ES) | 0.24257924 |
| Normalized Enrichment Score (NES) | 1.2513599 |
| Nominal p-value | 0.06966292 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAPKAPK2 | 45 | 4.126 | 0.0149 | Yes | ||
| 2 | FLNA | 73 | 3.845 | 0.0296 | Yes | ||
| 3 | ABL1 | 205 | 3.245 | 0.0361 | Yes | ||
| 4 | CSNK1E | 239 | 3.173 | 0.0476 | Yes | ||
| 5 | PPARG | 433 | 2.809 | 0.0489 | Yes | ||
| 6 | GRK6 | 578 | 2.636 | 0.0522 | Yes | ||
| 7 | TADA3L | 589 | 2.624 | 0.0626 | Yes | ||
| 8 | TGFB1I1 | 633 | 2.580 | 0.0711 | Yes | ||
| 9 | ARPC4 | 723 | 2.491 | 0.0768 | Yes | ||
| 10 | ASPH | 764 | 2.461 | 0.0849 | Yes | ||
| 11 | MYL6 | 780 | 2.450 | 0.0944 | Yes | ||
| 12 | SLC4A3 | 905 | 2.345 | 0.0975 | Yes | ||
| 13 | PDLIM7 | 909 | 2.343 | 0.1072 | Yes | ||
| 14 | NPPA | 968 | 2.303 | 0.1137 | Yes | ||
| 15 | BLOC1S1 | 1267 | 2.128 | 0.1065 | Yes | ||
| 16 | CTDSP1 | 1277 | 2.121 | 0.1149 | Yes | ||
| 17 | TITF1 | 1278 | 2.121 | 0.1238 | Yes | ||
| 18 | ACAA2 | 1310 | 2.106 | 0.1309 | Yes | ||
| 19 | MUS81 | 1397 | 2.071 | 0.1350 | Yes | ||
| 20 | DUSP10 | 1415 | 2.060 | 0.1427 | Yes | ||
| 21 | POU3F4 | 1447 | 2.043 | 0.1496 | Yes | ||
| 22 | SRD5A2 | 1473 | 2.034 | 0.1568 | Yes | ||
| 23 | CNN2 | 1513 | 2.020 | 0.1631 | Yes | ||
| 24 | GPR158 | 1796 | 1.907 | 0.1558 | Yes | ||
| 25 | ACTB | 1873 | 1.871 | 0.1596 | Yes | ||
| 26 | LRP5 | 1972 | 1.834 | 0.1620 | Yes | ||
| 27 | NPM3 | 1976 | 1.831 | 0.1695 | Yes | ||
| 28 | HIST1H4I | 1981 | 1.830 | 0.1769 | Yes | ||
| 29 | CX3CL1 | 2026 | 1.816 | 0.1822 | Yes | ||
| 30 | MRGPRF | 2067 | 1.800 | 0.1876 | Yes | ||
| 31 | EGR1 | 2211 | 1.746 | 0.1871 | Yes | ||
| 32 | LRRFIP1 | 2389 | 1.690 | 0.1846 | Yes | ||
| 33 | NR2F1 | 2420 | 1.681 | 0.1901 | Yes | ||
| 34 | CALD1 | 2423 | 1.681 | 0.1970 | Yes | ||
| 35 | EGR3 | 2449 | 1.673 | 0.2027 | Yes | ||
| 36 | CSMD3 | 2517 | 1.658 | 0.2060 | Yes | ||
| 37 | ITGB1BP2 | 2591 | 1.638 | 0.2089 | Yes | ||
| 38 | NAV1 | 2653 | 1.619 | 0.2124 | Yes | ||
| 39 | PRELP | 2706 | 1.602 | 0.2163 | Yes | ||
| 40 | TLE3 | 2721 | 1.599 | 0.2223 | Yes | ||
| 41 | LRRTM4 | 2875 | 1.554 | 0.2205 | Yes | ||
| 42 | ANXA6 | 3105 | 1.496 | 0.2143 | Yes | ||
| 43 | NR2F2 | 3482 | 1.401 | 0.1998 | Yes | ||
| 44 | PCDH7 | 3497 | 1.399 | 0.2049 | Yes | ||
| 45 | MAP1A | 3556 | 1.386 | 0.2076 | Yes | ||
| 46 | NR4A1 | 3620 | 1.372 | 0.2099 | Yes | ||
| 47 | COL1A1 | 3776 | 1.333 | 0.2071 | Yes | ||
| 48 | HOXC5 | 3861 | 1.315 | 0.2081 | Yes | ||
| 49 | DMD | 3883 | 1.309 | 0.2124 | Yes | ||
| 50 | RIMS1 | 3987 | 1.291 | 0.2123 | Yes | ||
| 51 | TAZ | 4089 | 1.272 | 0.2121 | Yes | ||
| 52 | ATP1A2 | 4168 | 1.252 | 0.2131 | Yes | ||
| 53 | CKM | 4214 | 1.244 | 0.2159 | Yes | ||
| 54 | FOXE3 | 4238 | 1.238 | 0.2199 | Yes | ||
| 55 | PODN | 4249 | 1.236 | 0.2245 | Yes | ||
| 56 | DVL3 | 4286 | 1.228 | 0.2277 | Yes | ||
| 57 | PFN1 | 4384 | 1.203 | 0.2275 | Yes | ||
| 58 | CDKN1B | 4465 | 1.185 | 0.2281 | Yes | ||
| 59 | MYLK | 4541 | 1.166 | 0.2290 | Yes | ||
| 60 | TPM2 | 4563 | 1.164 | 0.2327 | Yes | ||
| 61 | SCN3B | 4614 | 1.153 | 0.2348 | Yes | ||
| 62 | EPHB1 | 4636 | 1.148 | 0.2385 | Yes | ||
| 63 | PDLIM4 | 4710 | 1.134 | 0.2393 | Yes | ||
| 64 | ITGA7 | 4738 | 1.127 | 0.2426 | Yes | ||
| 65 | HOXB5 | 4898 | 1.098 | 0.2386 | No | ||
| 66 | STAT5B | 4945 | 1.090 | 0.2406 | No | ||
| 67 | FGF8 | 5109 | 1.059 | 0.2362 | No | ||
| 68 | MYO18B | 5375 | 1.007 | 0.2261 | No | ||
| 69 | HOXD13 | 5536 | 0.976 | 0.2215 | No | ||
| 70 | NOL4 | 5650 | 0.957 | 0.2194 | No | ||
| 71 | COL1A2 | 5779 | 0.932 | 0.2163 | No | ||
| 72 | SIX2 | 5780 | 0.932 | 0.2203 | No | ||
| 73 | RHOJ | 5881 | 0.914 | 0.2187 | No | ||
| 74 | SEPX1 | 5927 | 0.904 | 0.2200 | No | ||
| 75 | RARB | 6070 | 0.879 | 0.2160 | No | ||
| 76 | SIPA1 | 6109 | 0.872 | 0.2176 | No | ||
| 77 | TES | 6129 | 0.867 | 0.2202 | No | ||
| 78 | H2AFX | 6170 | 0.860 | 0.2217 | No | ||
| 79 | CITED2 | 6359 | 0.828 | 0.2149 | No | ||
| 80 | FGFRL1 | 6430 | 0.813 | 0.2145 | No | ||
| 81 | RUNDC1 | 6563 | 0.791 | 0.2107 | No | ||
| 82 | SRF | 6658 | 0.772 | 0.2088 | No | ||
| 83 | PLN | 6747 | 0.755 | 0.2072 | No | ||
| 84 | EMILIN2 | 6894 | 0.728 | 0.2024 | No | ||
| 85 | PRDM1 | 6917 | 0.726 | 0.2042 | No | ||
| 86 | LPP | 6929 | 0.723 | 0.2067 | No | ||
| 87 | MYADM | 6990 | 0.716 | 0.2064 | No | ||
| 88 | SLC7A1 | 7114 | 0.696 | 0.2027 | No | ||
| 89 | ACTG2 | 7160 | 0.687 | 0.2031 | No | ||
| 90 | WNT5A | 7213 | 0.677 | 0.2031 | No | ||
| 91 | NKX2-2 | 7314 | 0.661 | 0.2005 | No | ||
| 92 | RAI2 | 7478 | 0.629 | 0.1943 | No | ||
| 93 | CNN1 | 7538 | 0.619 | 0.1937 | No | ||
| 94 | RAP1A | 7716 | 0.583 | 0.1865 | No | ||
| 95 | JPH2 | 7725 | 0.582 | 0.1885 | No | ||
| 96 | GNG8 | 8067 | 0.518 | 0.1722 | No | ||
| 97 | FOS | 8087 | 0.514 | 0.1733 | No | ||
| 98 | ELAVL4 | 8121 | 0.507 | 0.1736 | No | ||
| 99 | HOXA1 | 8231 | 0.487 | 0.1698 | No | ||
| 100 | JUNB | 8312 | 0.471 | 0.1674 | No | ||
| 101 | MYO1E | 8342 | 0.466 | 0.1678 | No | ||
| 102 | FOSL1 | 8432 | 0.449 | 0.1648 | No | ||
| 103 | TNNC1 | 8808 | 0.381 | 0.1461 | No | ||
| 104 | AQP2 | 8856 | 0.372 | 0.1451 | No | ||
| 105 | ACTA1 | 9000 | 0.343 | 0.1388 | No | ||
| 106 | NKX6-1 | 9021 | 0.339 | 0.1391 | No | ||
| 107 | NFATC4 | 9023 | 0.339 | 0.1405 | No | ||
| 108 | TRDN | 9065 | 0.329 | 0.1396 | No | ||
| 109 | TAGLN | 9251 | 0.289 | 0.1308 | No | ||
| 110 | PLCB3 | 9357 | 0.267 | 0.1262 | No | ||
| 111 | DIXDC1 | 9365 | 0.266 | 0.1270 | No | ||
| 112 | PDLIM3 | 9369 | 0.265 | 0.1279 | No | ||
| 113 | RSU1 | 9483 | 0.237 | 0.1228 | No | ||
| 114 | NPAS2 | 9550 | 0.222 | 0.1201 | No | ||
| 115 | HOXD10 | 9760 | 0.177 | 0.1095 | No | ||
| 116 | HOXA5 | 9852 | 0.157 | 0.1053 | No | ||
| 117 | MSX1 | 9947 | 0.138 | 0.1007 | No | ||
| 118 | KCNMB1 | 10055 | 0.118 | 0.0954 | No | ||
| 119 | RASD2 | 10280 | 0.070 | 0.0836 | No | ||
| 120 | LDHA | 10361 | 0.050 | 0.0794 | No | ||
| 121 | LSP1 | 10362 | 0.050 | 0.0796 | No | ||
| 122 | KCNA1 | 10385 | 0.045 | 0.0786 | No | ||
| 123 | MYH11 | 10478 | 0.024 | 0.0737 | No | ||
| 124 | GADD45G | 11047 | -0.104 | 0.0433 | No | ||
| 125 | FST | 11104 | -0.114 | 0.0408 | No | ||
| 126 | IL17B | 11163 | -0.132 | 0.0382 | No | ||
| 127 | KCNK3 | 11349 | -0.171 | 0.0289 | No | ||
| 128 | SP7 | 11367 | -0.176 | 0.0287 | No | ||
| 129 | MYL9 | 11394 | -0.183 | 0.0280 | No | ||
| 130 | MAP2K6 | 11466 | -0.203 | 0.0250 | No | ||
| 131 | EGR2 | 11526 | -0.214 | 0.0227 | No | ||
| 132 | TRIM46 | 11715 | -0.254 | 0.0136 | No | ||
| 133 | NPR3 | 11744 | -0.262 | 0.0132 | No | ||
| 134 | CORO1C | 11788 | -0.273 | 0.0120 | No | ||
| 135 | INSM1 | 11988 | -0.322 | 0.0026 | No | ||
| 136 | MYL1 | 12236 | -0.379 | -0.0093 | No | ||
| 137 | TNMD | 12274 | -0.388 | -0.0096 | No | ||
| 138 | DUSP2 | 12418 | -0.425 | -0.0156 | No | ||
| 139 | CABP1 | 12480 | -0.442 | -0.0171 | No | ||
| 140 | FOXP1 | 12643 | -0.483 | -0.0238 | No | ||
| 141 | EFHD1 | 12748 | -0.508 | -0.0273 | No | ||
| 142 | VCL | 13115 | -0.607 | -0.0447 | No | ||
| 143 | KIF1B | 13122 | -0.610 | -0.0424 | No | ||
| 144 | ADAMTS12 | 13218 | -0.632 | -0.0449 | No | ||
| 145 | FOSB | 13389 | -0.687 | -0.0513 | No | ||
| 146 | THBS1 | 13426 | -0.697 | -0.0503 | No | ||
| 147 | RAB27A | 13698 | -0.780 | -0.0617 | No | ||
| 148 | STARD13 | 13801 | -0.811 | -0.0639 | No | ||
| 149 | PRKAB2 | 13834 | -0.818 | -0.0622 | No | ||
| 150 | IGF1 | 13921 | -0.840 | -0.0633 | No | ||
| 151 | BDNF | 14026 | -0.876 | -0.0653 | No | ||
| 152 | DGKG | 14124 | -0.910 | -0.0667 | No | ||
| 153 | ACTR3 | 14404 | -0.999 | -0.0777 | No | ||
| 154 | FOXP2 | 14497 | -1.024 | -0.0784 | No | ||
| 155 | CFL1 | 14556 | -1.041 | -0.0771 | No | ||
| 156 | RASSF2 | 14558 | -1.042 | -0.0728 | No | ||
| 157 | SLC25A4 | 14588 | -1.055 | -0.0700 | No | ||
| 158 | HOXA3 | 14738 | -1.112 | -0.0734 | No | ||
| 159 | KRTCAP2 | 14929 | -1.178 | -0.0787 | No | ||
| 160 | FOXP3 | 14998 | -1.207 | -0.0774 | No | ||
| 161 | PPAP2B | 15021 | -1.214 | -0.0735 | No | ||
| 162 | PHF12 | 15154 | -1.268 | -0.0753 | No | ||
| 163 | EML4 | 15341 | -1.339 | -0.0798 | No | ||
| 164 | GPR20 | 15377 | -1.352 | -0.0760 | No | ||
| 165 | DHH | 15505 | -1.407 | -0.0770 | No | ||
| 166 | SDPR | 15543 | -1.419 | -0.0730 | No | ||
| 167 | HIVEP3 | 15590 | -1.443 | -0.0695 | No | ||
| 168 | PICALM | 15769 | -1.519 | -0.0728 | No | ||
| 169 | ATF4 | 15863 | -1.564 | -0.0712 | No | ||
| 170 | NNAT | 16088 | -1.666 | -0.0764 | No | ||
| 171 | RERE | 16099 | -1.672 | -0.0699 | No | ||
| 172 | ABR | 16424 | -1.822 | -0.0799 | No | ||
| 173 | H3F3A | 16494 | -1.854 | -0.0758 | No | ||
| 174 | RRM1 | 16502 | -1.857 | -0.0684 | No | ||
| 175 | MYL7 | 16619 | -1.921 | -0.0666 | No | ||
| 176 | CACNA1B | 16685 | -1.951 | -0.0620 | No | ||
| 177 | MATR3 | 16687 | -1.952 | -0.0538 | No | ||
| 178 | CFL2 | 16783 | -2.009 | -0.0506 | No | ||
| 179 | PPP1R12A | 17014 | -2.128 | -0.0541 | No | ||
| 180 | UBE2D3 | 17090 | -2.167 | -0.0491 | No | ||
| 181 | SYNCRIP | 17454 | -2.383 | -0.0588 | No | ||
| 182 | PADI2 | 17545 | -2.437 | -0.0534 | No | ||
| 183 | SCOC | 17786 | -2.594 | -0.0556 | No | ||
| 184 | MBNL1 | 17818 | -2.617 | -0.0463 | No | ||
| 185 | RBBP7 | 17920 | -2.697 | -0.0404 | No | ||
| 186 | SVIL | 18029 | -2.790 | -0.0346 | No | ||
| 187 | SIN3A | 18050 | -2.813 | -0.0238 | No | ||
| 188 | TPM3 | 18287 | -3.115 | -0.0236 | No | ||
| 189 | CNTF | 18293 | -3.120 | -0.0108 | No | ||
| 190 | ROCK2 | 18399 | -3.300 | -0.0026 | No | ||
| 191 | HOXB4 | 18436 | -3.412 | 0.0098 | No |
