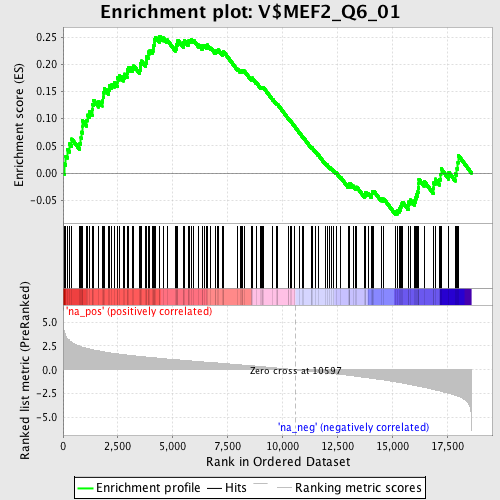
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MEF2_Q6_01 |
| Enrichment Score (ES) | 0.25173426 |
| Normalized Enrichment Score (NES) | 1.2612171 |
| Nominal p-value | 0.055666003 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 48 | 4.113 | 0.0180 | Yes | ||
| 2 | RASGEF1B | 123 | 3.482 | 0.0314 | Yes | ||
| 3 | IL10 | 195 | 3.274 | 0.0439 | Yes | ||
| 4 | GPR30 | 294 | 3.054 | 0.0539 | Yes | ||
| 5 | MAP2K5 | 395 | 2.871 | 0.0628 | Yes | ||
| 6 | ASPH | 764 | 2.461 | 0.0552 | Yes | ||
| 7 | ALDOA | 812 | 2.426 | 0.0648 | Yes | ||
| 8 | DCTN1 | 835 | 2.405 | 0.0756 | Yes | ||
| 9 | B4GALT1 | 877 | 2.372 | 0.0852 | Yes | ||
| 10 | ART3 | 883 | 2.367 | 0.0968 | Yes | ||
| 11 | CAMK1 | 1079 | 2.238 | 0.0974 | Yes | ||
| 12 | PER1 | 1111 | 2.216 | 0.1068 | Yes | ||
| 13 | ALS2CR2 | 1201 | 2.166 | 0.1128 | Yes | ||
| 14 | RFX3 | 1324 | 2.101 | 0.1167 | Yes | ||
| 15 | SLC26A9 | 1336 | 2.095 | 0.1266 | Yes | ||
| 16 | KTN1 | 1386 | 2.074 | 0.1343 | Yes | ||
| 17 | PPP2R2A | 1627 | 1.973 | 0.1312 | Yes | ||
| 18 | LBX1 | 1775 | 1.915 | 0.1328 | Yes | ||
| 19 | RASGRF1 | 1818 | 1.900 | 0.1400 | Yes | ||
| 20 | EEF1A2 | 1842 | 1.888 | 0.1482 | Yes | ||
| 21 | GATA4 | 1866 | 1.875 | 0.1563 | Yes | ||
| 22 | USP2 | 2084 | 1.793 | 0.1536 | Yes | ||
| 23 | CACNA2D3 | 2116 | 1.782 | 0.1608 | Yes | ||
| 24 | TBR1 | 2224 | 1.742 | 0.1637 | Yes | ||
| 25 | CLCN1 | 2330 | 1.705 | 0.1665 | Yes | ||
| 26 | HIST1H2BA | 2474 | 1.667 | 0.1671 | Yes | ||
| 27 | CAPN3 | 2479 | 1.666 | 0.1752 | Yes | ||
| 28 | HFE2 | 2559 | 1.645 | 0.1792 | Yes | ||
| 29 | PI15 | 2745 | 1.592 | 0.1771 | Yes | ||
| 30 | TEF | 2799 | 1.577 | 0.1821 | Yes | ||
| 31 | RIMS2 | 2918 | 1.543 | 0.1835 | Yes | ||
| 32 | INPPL1 | 2937 | 1.538 | 0.1902 | Yes | ||
| 33 | LIF | 2994 | 1.523 | 0.1948 | Yes | ||
| 34 | MFGE8 | 3154 | 1.480 | 0.1935 | Yes | ||
| 35 | SOX5 | 3225 | 1.461 | 0.1971 | Yes | ||
| 36 | MYOZ2 | 3500 | 1.398 | 0.1892 | Yes | ||
| 37 | SMPX | 3518 | 1.394 | 0.1952 | Yes | ||
| 38 | TYRO3 | 3535 | 1.390 | 0.2013 | Yes | ||
| 39 | ACCN1 | 3574 | 1.381 | 0.2062 | Yes | ||
| 40 | COL1A1 | 3776 | 1.333 | 0.2019 | Yes | ||
| 41 | GRB14 | 3795 | 1.330 | 0.2076 | Yes | ||
| 42 | PACS1 | 3804 | 1.328 | 0.2138 | Yes | ||
| 43 | HDAC7A | 3879 | 1.310 | 0.2164 | Yes | ||
| 44 | DMD | 3883 | 1.309 | 0.2228 | Yes | ||
| 45 | CRTAP | 3957 | 1.296 | 0.2253 | Yes | ||
| 46 | TCEA3 | 4060 | 1.276 | 0.2261 | Yes | ||
| 47 | INPP4A | 4101 | 1.269 | 0.2303 | Yes | ||
| 48 | MYH6 | 4133 | 1.260 | 0.2349 | Yes | ||
| 49 | ADCY6 | 4158 | 1.254 | 0.2399 | Yes | ||
| 50 | LRRC20 | 4163 | 1.253 | 0.2460 | Yes | ||
| 51 | CKM | 4214 | 1.244 | 0.2495 | Yes | ||
| 52 | MYOG | 4381 | 1.204 | 0.2465 | Yes | ||
| 53 | PABPC4 | 4396 | 1.200 | 0.2517 | Yes | ||
| 54 | TPM2 | 4563 | 1.164 | 0.2486 | No | ||
| 55 | ITGA7 | 4738 | 1.127 | 0.2448 | No | ||
| 56 | TNNC2 | 5120 | 1.056 | 0.2294 | No | ||
| 57 | CASQ1 | 5167 | 1.047 | 0.2321 | No | ||
| 58 | ETV1 | 5172 | 1.046 | 0.2372 | No | ||
| 59 | NEDD4 | 5216 | 1.039 | 0.2400 | No | ||
| 60 | HIST1H1T | 5230 | 1.036 | 0.2445 | No | ||
| 61 | RALY | 5484 | 0.985 | 0.2357 | No | ||
| 62 | SNAP25 | 5493 | 0.983 | 0.2402 | No | ||
| 63 | DIRAS1 | 5514 | 0.980 | 0.2440 | No | ||
| 64 | KBTBD5 | 5699 | 0.947 | 0.2388 | No | ||
| 65 | PRX | 5710 | 0.946 | 0.2430 | No | ||
| 66 | ZFPM2 | 5771 | 0.934 | 0.2444 | No | ||
| 67 | POFUT1 | 5840 | 0.921 | 0.2453 | No | ||
| 68 | BNC2 | 5950 | 0.900 | 0.2439 | No | ||
| 69 | P2RX5 | 6182 | 0.858 | 0.2357 | No | ||
| 70 | KCNA7 | 6338 | 0.831 | 0.2314 | No | ||
| 71 | TRHR | 6365 | 0.827 | 0.2342 | No | ||
| 72 | MID2 | 6431 | 0.813 | 0.2347 | No | ||
| 73 | AQP1 | 6532 | 0.797 | 0.2333 | No | ||
| 74 | HSPB3 | 6562 | 0.791 | 0.2356 | No | ||
| 75 | KCNN1 | 6736 | 0.757 | 0.2301 | No | ||
| 76 | TPBG | 6943 | 0.722 | 0.2225 | No | ||
| 77 | DLL4 | 6967 | 0.719 | 0.2249 | No | ||
| 78 | SSPN | 7034 | 0.708 | 0.2248 | No | ||
| 79 | RHOB | 7066 | 0.704 | 0.2267 | No | ||
| 80 | CMYA1 | 7264 | 0.669 | 0.2193 | No | ||
| 81 | MYO9A | 7300 | 0.665 | 0.2208 | No | ||
| 82 | JUN | 7319 | 0.660 | 0.2231 | No | ||
| 83 | KCNJ9 | 7954 | 0.538 | 0.1914 | No | ||
| 84 | DNAJA4 | 8088 | 0.514 | 0.1868 | No | ||
| 85 | ELAVL4 | 8121 | 0.507 | 0.1876 | No | ||
| 86 | SV2A | 8154 | 0.500 | 0.1883 | No | ||
| 87 | IAPP | 8195 | 0.493 | 0.1886 | No | ||
| 88 | MAFA | 8256 | 0.482 | 0.1878 | No | ||
| 89 | FGF13 | 8577 | 0.420 | 0.1726 | No | ||
| 90 | MYL2 | 8600 | 0.416 | 0.1734 | No | ||
| 91 | PCDH9 | 8611 | 0.414 | 0.1750 | No | ||
| 92 | TNNC1 | 8808 | 0.381 | 0.1663 | No | ||
| 93 | ADAM11 | 9011 | 0.341 | 0.1570 | No | ||
| 94 | TMEPAI | 9033 | 0.337 | 0.1576 | No | ||
| 95 | TRDN | 9065 | 0.329 | 0.1575 | No | ||
| 96 | IGJ | 9098 | 0.323 | 0.1574 | No | ||
| 97 | SLC16A13 | 9121 | 0.318 | 0.1578 | No | ||
| 98 | ESR1 | 9562 | 0.220 | 0.1351 | No | ||
| 99 | TNNI3K | 9718 | 0.185 | 0.1276 | No | ||
| 100 | CKMT2 | 9754 | 0.178 | 0.1266 | No | ||
| 101 | UBXD3 | 9775 | 0.172 | 0.1264 | No | ||
| 102 | PDGFRA | 10254 | 0.075 | 0.1008 | No | ||
| 103 | CPT1B | 10287 | 0.068 | 0.0994 | No | ||
| 104 | SMARCA1 | 10344 | 0.053 | 0.0967 | No | ||
| 105 | CLDN14 | 10409 | 0.039 | 0.0934 | No | ||
| 106 | RARA | 10544 | 0.010 | 0.0862 | No | ||
| 107 | LECT1 | 10792 | -0.049 | 0.0730 | No | ||
| 108 | BZW2 | 10925 | -0.078 | 0.0663 | No | ||
| 109 | COL8A1 | 10976 | -0.088 | 0.0640 | No | ||
| 110 | SCN3A | 11303 | -0.161 | 0.0471 | No | ||
| 111 | LRRC2 | 11326 | -0.166 | 0.0468 | No | ||
| 112 | USP13 | 11347 | -0.171 | 0.0465 | No | ||
| 113 | GPR27 | 11485 | -0.207 | 0.0402 | No | ||
| 114 | SLC2A4 | 11623 | -0.234 | 0.0339 | No | ||
| 115 | NOG | 11937 | -0.308 | 0.0185 | No | ||
| 116 | NR0B2 | 12027 | -0.331 | 0.0153 | No | ||
| 117 | HIPK1 | 12159 | -0.362 | 0.0100 | No | ||
| 118 | MYL1 | 12236 | -0.379 | 0.0078 | No | ||
| 119 | KPNA3 | 12330 | -0.403 | 0.0048 | No | ||
| 120 | STAC | 12440 | -0.429 | 0.0010 | No | ||
| 121 | FOXP1 | 12643 | -0.483 | -0.0075 | No | ||
| 122 | RCOR1 | 13002 | -0.575 | -0.0240 | No | ||
| 123 | SLC8A3 | 13050 | -0.589 | -0.0236 | No | ||
| 124 | HIST1H2AA | 13056 | -0.592 | -0.0210 | No | ||
| 125 | ART5 | 13064 | -0.593 | -0.0184 | No | ||
| 126 | ADAMTS12 | 13218 | -0.632 | -0.0235 | No | ||
| 127 | HIPK3 | 13334 | -0.671 | -0.0264 | No | ||
| 128 | MITF | 13381 | -0.686 | -0.0254 | No | ||
| 129 | WFDC1 | 13746 | -0.798 | -0.0412 | No | ||
| 130 | HS6ST2 | 13767 | -0.803 | -0.0382 | No | ||
| 131 | EPHA7 | 13784 | -0.807 | -0.0351 | No | ||
| 132 | DPP10 | 13906 | -0.836 | -0.0375 | No | ||
| 133 | TFB2M | 14045 | -0.881 | -0.0405 | No | ||
| 134 | PPARGC1A | 14078 | -0.893 | -0.0378 | No | ||
| 135 | TNNT2 | 14087 | -0.896 | -0.0338 | No | ||
| 136 | CNTN1 | 14158 | -0.919 | -0.0330 | No | ||
| 137 | FOXP2 | 14497 | -1.024 | -0.0461 | No | ||
| 138 | LYN | 14620 | -1.067 | -0.0474 | No | ||
| 139 | CASK | 15148 | -1.265 | -0.0697 | No | ||
| 140 | ANGPT2 | 15252 | -1.309 | -0.0687 | No | ||
| 141 | PLAGL2 | 15314 | -1.330 | -0.0653 | No | ||
| 142 | PPP1R16B | 15374 | -1.351 | -0.0618 | No | ||
| 143 | FGF12 | 15404 | -1.365 | -0.0565 | No | ||
| 144 | RHOBTB1 | 15475 | -1.396 | -0.0534 | No | ||
| 145 | SIX1 | 15736 | -1.507 | -0.0599 | No | ||
| 146 | CASQ2 | 15747 | -1.511 | -0.0529 | No | ||
| 147 | PPM1B | 15820 | -1.541 | -0.0491 | No | ||
| 148 | SLC6A13 | 16004 | -1.629 | -0.0509 | No | ||
| 149 | SLC26A6 | 16056 | -1.653 | -0.0454 | No | ||
| 150 | RERE | 16099 | -1.672 | -0.0393 | No | ||
| 151 | GABARAPL2 | 16167 | -1.699 | -0.0344 | No | ||
| 152 | ATP1B2 | 16186 | -1.708 | -0.0268 | No | ||
| 153 | CTBP2 | 16208 | -1.718 | -0.0194 | No | ||
| 154 | CDC42EP3 | 16215 | -1.721 | -0.0111 | No | ||
| 155 | GNB4 | 16452 | -1.832 | -0.0147 | No | ||
| 156 | ZADH2 | 16874 | -2.055 | -0.0273 | No | ||
| 157 | COL13A1 | 16892 | -2.063 | -0.0179 | No | ||
| 158 | CUTL1 | 16954 | -2.094 | -0.0107 | No | ||
| 159 | MBNL2 | 17164 | -2.211 | -0.0110 | No | ||
| 160 | RAP2C | 17200 | -2.232 | -0.0017 | No | ||
| 161 | TPP2 | 17224 | -2.244 | 0.0083 | No | ||
| 162 | TRIM33 | 17582 | -2.459 | 0.0012 | No | ||
| 163 | NLK | 17875 | -2.665 | -0.0013 | No | ||
| 164 | WBP4 | 17948 | -2.716 | 0.0084 | No | ||
| 165 | ANKMY2 | 17993 | -2.757 | 0.0198 | No | ||
| 166 | AMMECR1 | 18018 | -2.779 | 0.0324 | No |
