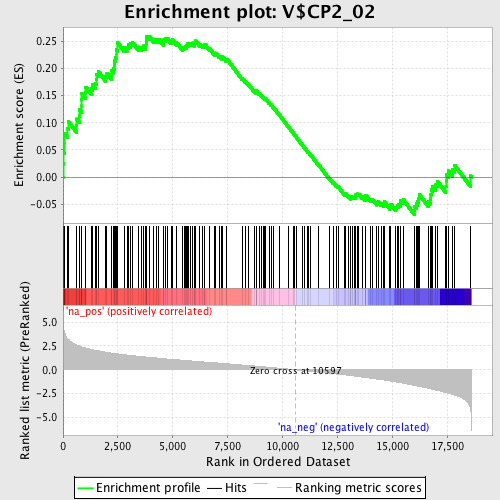
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$CP2_02 |
| Enrichment Score (ES) | 0.2592479 |
| Normalized Enrichment Score (NES) | 1.2974067 |
| Nominal p-value | 0.039748956 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UBE1 | 4 | 5.353 | 0.0257 | Yes | ||
| 2 | FLOT2 | 35 | 4.221 | 0.0444 | Yes | ||
| 3 | GNAQ | 64 | 3.955 | 0.0620 | Yes | ||
| 4 | BTBD14B | 68 | 3.904 | 0.0807 | Yes | ||
| 5 | HTATIP | 206 | 3.244 | 0.0890 | Yes | ||
| 6 | NRXN3 | 240 | 3.171 | 0.1025 | Yes | ||
| 7 | PTCRA | 615 | 2.596 | 0.0948 | Yes | ||
| 8 | MNT | 624 | 2.589 | 0.1069 | Yes | ||
| 9 | MAPK3 | 736 | 2.478 | 0.1129 | Yes | ||
| 10 | PRKCH | 756 | 2.466 | 0.1238 | Yes | ||
| 11 | ALDH3B1 | 840 | 2.401 | 0.1309 | Yes | ||
| 12 | RHBDF1 | 844 | 2.396 | 0.1423 | Yes | ||
| 13 | CREB1 | 854 | 2.389 | 0.1533 | Yes | ||
| 14 | PTHLH | 1011 | 2.275 | 0.1559 | Yes | ||
| 15 | PRKAG1 | 1035 | 2.261 | 0.1656 | Yes | ||
| 16 | NEK8 | 1281 | 2.120 | 0.1625 | Yes | ||
| 17 | SLC26A9 | 1336 | 2.095 | 0.1697 | Yes | ||
| 18 | AQP6 | 1466 | 2.036 | 0.1726 | Yes | ||
| 19 | KCNE3 | 1512 | 2.021 | 0.1799 | Yes | ||
| 20 | TUFT1 | 1531 | 2.012 | 0.1887 | Yes | ||
| 21 | EFNA3 | 1609 | 1.979 | 0.1941 | Yes | ||
| 22 | TGFBR2 | 1942 | 1.846 | 0.1850 | Yes | ||
| 23 | ADRA1A | 1996 | 1.825 | 0.1909 | Yes | ||
| 24 | CLDN4 | 2208 | 1.748 | 0.1880 | Yes | ||
| 25 | CRK | 2219 | 1.743 | 0.1958 | Yes | ||
| 26 | PCDHB4 | 2304 | 1.715 | 0.1996 | Yes | ||
| 27 | DUSP6 | 2326 | 1.706 | 0.2067 | Yes | ||
| 28 | BRUNOL6 | 2346 | 1.701 | 0.2139 | Yes | ||
| 29 | PAK7 | 2378 | 1.693 | 0.2204 | Yes | ||
| 30 | NR2F1 | 2420 | 1.681 | 0.2263 | Yes | ||
| 31 | CALD1 | 2423 | 1.681 | 0.2343 | Yes | ||
| 32 | LDB2 | 2481 | 1.666 | 0.2393 | Yes | ||
| 33 | MAST2 | 2482 | 1.665 | 0.2473 | Yes | ||
| 34 | KLHDC3 | 2791 | 1.579 | 0.2383 | Yes | ||
| 35 | FRMD3 | 2914 | 1.543 | 0.2391 | Yes | ||
| 36 | HPCAL1 | 2964 | 1.532 | 0.2439 | Yes | ||
| 37 | XPNPEP1 | 3059 | 1.506 | 0.2460 | Yes | ||
| 38 | NR1D1 | 3151 | 1.482 | 0.2483 | Yes | ||
| 39 | PAPPA | 3444 | 1.409 | 0.2393 | Yes | ||
| 40 | EPN3 | 3580 | 1.380 | 0.2386 | Yes | ||
| 41 | MRPL14 | 3655 | 1.364 | 0.2412 | Yes | ||
| 42 | LHX2 | 3769 | 1.335 | 0.2415 | Yes | ||
| 43 | KIF1C | 3778 | 1.333 | 0.2475 | Yes | ||
| 44 | TLN1 | 3800 | 1.329 | 0.2528 | Yes | ||
| 45 | KCTD15 | 3809 | 1.328 | 0.2588 | Yes | ||
| 46 | P518 | 3918 | 1.303 | 0.2592 | Yes | ||
| 47 | FEV | 4131 | 1.261 | 0.2539 | No | ||
| 48 | PODN | 4249 | 1.236 | 0.2535 | No | ||
| 49 | PRKACA | 4367 | 1.207 | 0.2530 | No | ||
| 50 | FOXA1 | 4580 | 1.160 | 0.2471 | No | ||
| 51 | HSPA1L | 4589 | 1.158 | 0.2523 | No | ||
| 52 | TXNL5 | 4650 | 1.147 | 0.2546 | No | ||
| 53 | RASSF1 | 4735 | 1.128 | 0.2555 | No | ||
| 54 | ZNF385 | 4930 | 1.092 | 0.2502 | No | ||
| 55 | CAPN6 | 4965 | 1.087 | 0.2536 | No | ||
| 56 | ETV1 | 5172 | 1.046 | 0.2475 | No | ||
| 57 | GMPPB | 5438 | 0.996 | 0.2380 | No | ||
| 58 | DSC2 | 5512 | 0.981 | 0.2388 | No | ||
| 59 | RNF121 | 5566 | 0.970 | 0.2406 | No | ||
| 60 | TRAF4 | 5628 | 0.959 | 0.2419 | No | ||
| 61 | NOL4 | 5650 | 0.957 | 0.2454 | No | ||
| 62 | FGF9 | 5732 | 0.942 | 0.2456 | No | ||
| 63 | CITED1 | 5808 | 0.927 | 0.2460 | No | ||
| 64 | ARX | 5879 | 0.915 | 0.2466 | No | ||
| 65 | TRERF1 | 5987 | 0.894 | 0.2452 | No | ||
| 66 | KIF5A | 6009 | 0.891 | 0.2483 | No | ||
| 67 | GABARAP | 6049 | 0.884 | 0.2505 | No | ||
| 68 | HIF1A | 6233 | 0.849 | 0.2447 | No | ||
| 69 | PPP2R5C | 6354 | 0.829 | 0.2422 | No | ||
| 70 | MID2 | 6431 | 0.813 | 0.2420 | No | ||
| 71 | MOGAT2 | 6459 | 0.809 | 0.2444 | No | ||
| 72 | DDR1 | 6676 | 0.769 | 0.2364 | No | ||
| 73 | ODC1 | 6913 | 0.727 | 0.2272 | No | ||
| 74 | CAP1 | 6958 | 0.720 | 0.2283 | No | ||
| 75 | SRMS | 7126 | 0.694 | 0.2226 | No | ||
| 76 | C1QTNF5 | 7236 | 0.673 | 0.2199 | No | ||
| 77 | BACH1 | 7275 | 0.667 | 0.2211 | No | ||
| 78 | DEF6 | 7434 | 0.639 | 0.2156 | No | ||
| 79 | IRX3 | 7464 | 0.632 | 0.2171 | No | ||
| 80 | GRB7 | 8168 | 0.497 | 0.1814 | No | ||
| 81 | JUNB | 8312 | 0.471 | 0.1759 | No | ||
| 82 | RYR1 | 8460 | 0.443 | 0.1701 | No | ||
| 83 | CAMK2G | 8727 | 0.394 | 0.1576 | No | ||
| 84 | MAPRE3 | 8799 | 0.382 | 0.1556 | No | ||
| 85 | TNNC1 | 8808 | 0.381 | 0.1570 | No | ||
| 86 | CNTN6 | 8812 | 0.381 | 0.1587 | No | ||
| 87 | ARHGAP5 | 8931 | 0.356 | 0.1540 | No | ||
| 88 | NFATC4 | 9023 | 0.339 | 0.1507 | No | ||
| 89 | DDIT4 | 9125 | 0.317 | 0.1468 | No | ||
| 90 | STAC2 | 9200 | 0.297 | 0.1442 | No | ||
| 91 | DCX | 9246 | 0.290 | 0.1432 | No | ||
| 92 | GSH1 | 9392 | 0.258 | 0.1366 | No | ||
| 93 | PPP1R9B | 9478 | 0.238 | 0.1331 | No | ||
| 94 | PLXNA3 | 9599 | 0.212 | 0.1276 | No | ||
| 95 | SLC9A1 | 9848 | 0.158 | 0.1150 | No | ||
| 96 | SLC2A9 | 10274 | 0.071 | 0.0923 | No | ||
| 97 | RHBDL1 | 10477 | 0.024 | 0.0815 | No | ||
| 98 | DDAH2 | 10541 | 0.012 | 0.0781 | No | ||
| 99 | TRPV2 | 10619 | -0.005 | 0.0739 | No | ||
| 100 | TBX2 | 10920 | -0.078 | 0.0581 | No | ||
| 101 | GPC3 | 10980 | -0.089 | 0.0553 | No | ||
| 102 | TLX3 | 11159 | -0.130 | 0.0463 | No | ||
| 103 | FBS1 | 11189 | -0.137 | 0.0454 | No | ||
| 104 | PHPT1 | 11271 | -0.154 | 0.0417 | No | ||
| 105 | GNB3 | 11644 | -0.238 | 0.0227 | No | ||
| 106 | SLC25A28 | 12151 | -0.360 | -0.0030 | No | ||
| 107 | NRF1 | 12325 | -0.402 | -0.0104 | No | ||
| 108 | RPL41 | 12461 | -0.436 | -0.0156 | No | ||
| 109 | CS | 12539 | -0.458 | -0.0176 | No | ||
| 110 | FKBP2 | 12830 | -0.528 | -0.0307 | No | ||
| 111 | BMP4 | 12869 | -0.538 | -0.0302 | No | ||
| 112 | NGFRAP1 | 13004 | -0.576 | -0.0346 | No | ||
| 113 | COX8C | 13111 | -0.606 | -0.0375 | No | ||
| 114 | WNT10B | 13116 | -0.607 | -0.0347 | No | ||
| 115 | UPK2 | 13208 | -0.631 | -0.0366 | No | ||
| 116 | SLC25A14 | 13271 | -0.650 | -0.0368 | No | ||
| 117 | KIFAP3 | 13313 | -0.665 | -0.0359 | No | ||
| 118 | PAX2 | 13324 | -0.668 | -0.0332 | No | ||
| 119 | LZTS2 | 13346 | -0.677 | -0.0310 | No | ||
| 120 | TBC1D5 | 13404 | -0.692 | -0.0308 | No | ||
| 121 | GLTP | 13474 | -0.709 | -0.0311 | No | ||
| 122 | HOXB6 | 13625 | -0.760 | -0.0355 | No | ||
| 123 | RTN1 | 13768 | -0.803 | -0.0393 | No | ||
| 124 | MMP11 | 13781 | -0.806 | -0.0361 | No | ||
| 125 | STARD13 | 13801 | -0.811 | -0.0332 | No | ||
| 126 | BDNF | 14026 | -0.876 | -0.0411 | No | ||
| 127 | DSCAM | 14101 | -0.903 | -0.0408 | No | ||
| 128 | SHB | 14297 | -0.963 | -0.0467 | No | ||
| 129 | PITX2 | 14353 | -0.981 | -0.0449 | No | ||
| 130 | FOXP2 | 14497 | -1.024 | -0.0477 | No | ||
| 131 | LYN | 14620 | -1.067 | -0.0492 | No | ||
| 132 | NISCH | 14641 | -1.074 | -0.0450 | No | ||
| 133 | PFTK1 | 14897 | -1.166 | -0.0532 | No | ||
| 134 | RAB10 | 14942 | -1.183 | -0.0499 | No | ||
| 135 | SCAMP5 | 15149 | -1.266 | -0.0549 | No | ||
| 136 | KCNH2 | 15217 | -1.293 | -0.0523 | No | ||
| 137 | NRIP2 | 15283 | -1.320 | -0.0495 | No | ||
| 138 | PPP1R16B | 15374 | -1.351 | -0.0478 | No | ||
| 139 | GSK3B | 15391 | -1.361 | -0.0421 | No | ||
| 140 | DHH | 15505 | -1.407 | -0.0414 | No | ||
| 141 | FLI1 | 16008 | -1.630 | -0.0607 | No | ||
| 142 | MDK | 16022 | -1.639 | -0.0535 | No | ||
| 143 | NNAT | 16088 | -1.666 | -0.0490 | No | ||
| 144 | CPNE1 | 16149 | -1.691 | -0.0441 | No | ||
| 145 | CDC42EP3 | 16215 | -1.721 | -0.0393 | No | ||
| 146 | GPRC5C | 16225 | -1.725 | -0.0314 | No | ||
| 147 | ATP10D | 16643 | -1.931 | -0.0447 | No | ||
| 148 | YWHAG | 16732 | -1.980 | -0.0399 | No | ||
| 149 | BCL11B | 16759 | -1.998 | -0.0316 | No | ||
| 150 | LTBP1 | 16784 | -2.009 | -0.0232 | No | ||
| 151 | DNAJB1 | 16848 | -2.040 | -0.0168 | No | ||
| 152 | CHD4 | 16990 | -2.116 | -0.0142 | No | ||
| 153 | RBBP6 | 17062 | -2.151 | -0.0076 | No | ||
| 154 | MORF4L2 | 17449 | -2.380 | -0.0170 | No | ||
| 155 | NTRK3 | 17459 | -2.384 | -0.0060 | No | ||
| 156 | FOXL2 | 17484 | -2.400 | 0.0043 | No | ||
| 157 | HCFC1 | 17565 | -2.449 | 0.0118 | No | ||
| 158 | H3F3B | 17752 | -2.572 | 0.0141 | No | ||
| 159 | TRIM8 | 17841 | -2.636 | 0.0221 | No | ||
| 160 | EPC1 | 18571 | -4.100 | 0.0024 | No |
