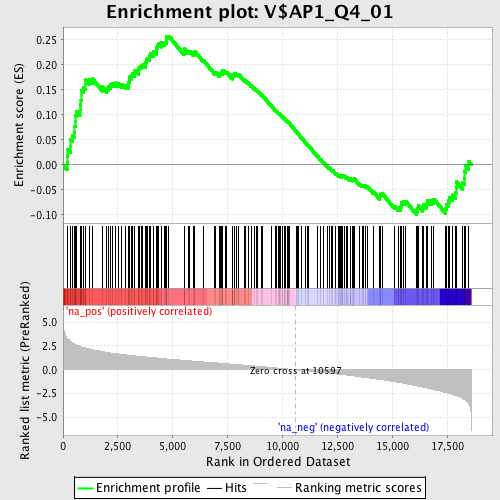
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$AP1_Q4_01 |
| Enrichment Score (ES) | 0.25752863 |
| Normalized Enrichment Score (NES) | 1.3102291 |
| Nominal p-value | 0.030237582 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ETV5 | 182 | 3.305 | 0.0047 | Yes | ||
| 2 | IL10 | 195 | 3.274 | 0.0186 | Yes | ||
| 3 | TYSND1 | 215 | 3.223 | 0.0318 | Yes | ||
| 4 | ENO1 | 349 | 2.947 | 0.0376 | Yes | ||
| 5 | MARK4 | 352 | 2.944 | 0.0505 | Yes | ||
| 6 | RB1CC1 | 421 | 2.822 | 0.0593 | Yes | ||
| 7 | VASP | 522 | 2.698 | 0.0658 | Yes | ||
| 8 | ROM1 | 534 | 2.685 | 0.0771 | Yes | ||
| 9 | MAP4K5 | 556 | 2.658 | 0.0877 | Yes | ||
| 10 | SQSTM1 | 564 | 2.652 | 0.0991 | Yes | ||
| 11 | MNT | 624 | 2.589 | 0.1073 | Yes | ||
| 12 | CDH23 | 770 | 2.460 | 0.1103 | Yes | ||
| 13 | NCDN | 773 | 2.457 | 0.1211 | Yes | ||
| 14 | ALDOA | 812 | 2.426 | 0.1297 | Yes | ||
| 15 | RHBDF1 | 844 | 2.396 | 0.1386 | Yes | ||
| 16 | P2RXL1 | 846 | 2.396 | 0.1492 | Yes | ||
| 17 | SLC35B1 | 930 | 2.330 | 0.1550 | Yes | ||
| 18 | TUBB4 | 1007 | 2.279 | 0.1609 | Yes | ||
| 19 | CSF3 | 1008 | 2.276 | 0.1710 | Yes | ||
| 20 | ITGB4 | 1191 | 2.168 | 0.1707 | Yes | ||
| 21 | SLC26A9 | 1336 | 2.095 | 0.1722 | Yes | ||
| 22 | CD68 | 1788 | 1.910 | 0.1561 | Yes | ||
| 23 | AQP5 | 1997 | 1.824 | 0.1529 | Yes | ||
| 24 | USP2 | 2084 | 1.793 | 0.1562 | Yes | ||
| 25 | CHST4 | 2158 | 1.766 | 0.1600 | Yes | ||
| 26 | PEA15 | 2235 | 1.738 | 0.1636 | Yes | ||
| 27 | ITPKC | 2365 | 1.697 | 0.1641 | Yes | ||
| 28 | CSMD3 | 2517 | 1.658 | 0.1632 | Yes | ||
| 29 | BAG2 | 2682 | 1.610 | 0.1615 | Yes | ||
| 30 | VIT | 2844 | 1.565 | 0.1596 | Yes | ||
| 31 | ELK3 | 2974 | 1.529 | 0.1594 | Yes | ||
| 32 | SEC24D | 2987 | 1.525 | 0.1655 | Yes | ||
| 33 | TREX1 | 3018 | 1.517 | 0.1706 | Yes | ||
| 34 | ECM1 | 3044 | 1.510 | 0.1759 | Yes | ||
| 35 | DUSP14 | 3124 | 1.491 | 0.1782 | Yes | ||
| 36 | NR1D1 | 3151 | 1.482 | 0.1834 | Yes | ||
| 37 | HSPB8 | 3271 | 1.450 | 0.1833 | Yes | ||
| 38 | RPS6KA4 | 3274 | 1.450 | 0.1896 | Yes | ||
| 39 | FBXO44 | 3427 | 1.412 | 0.1876 | Yes | ||
| 40 | PAPPA | 3444 | 1.409 | 0.1930 | Yes | ||
| 41 | MYOZ2 | 3500 | 1.398 | 0.1962 | Yes | ||
| 42 | EPHB2 | 3570 | 1.382 | 0.1985 | Yes | ||
| 43 | WDFY3 | 3640 | 1.369 | 0.2008 | Yes | ||
| 44 | RIT1 | 3744 | 1.341 | 0.2012 | Yes | ||
| 45 | HDAC3 | 3770 | 1.334 | 0.2057 | Yes | ||
| 46 | SV2B | 3782 | 1.332 | 0.2110 | Yes | ||
| 47 | ADCK4 | 3840 | 1.321 | 0.2138 | Yes | ||
| 48 | SPATS2 | 3943 | 1.299 | 0.2140 | Yes | ||
| 49 | ADCY8 | 3948 | 1.297 | 0.2195 | Yes | ||
| 50 | RIMS1 | 3987 | 1.291 | 0.2231 | Yes | ||
| 51 | MPV17 | 4119 | 1.264 | 0.2216 | Yes | ||
| 52 | COL16A1 | 4132 | 1.260 | 0.2265 | Yes | ||
| 53 | ITM2B | 4245 | 1.238 | 0.2259 | Yes | ||
| 54 | PADI4 | 4262 | 1.233 | 0.2305 | Yes | ||
| 55 | PSTPIP1 | 4270 | 1.231 | 0.2356 | Yes | ||
| 56 | CSF1R | 4307 | 1.222 | 0.2390 | Yes | ||
| 57 | SFN | 4343 | 1.213 | 0.2425 | Yes | ||
| 58 | ELAVL2 | 4470 | 1.184 | 0.2409 | Yes | ||
| 59 | MARK1 | 4488 | 1.180 | 0.2452 | Yes | ||
| 60 | OLR1 | 4613 | 1.153 | 0.2436 | Yes | ||
| 61 | LTBP3 | 4682 | 1.140 | 0.2449 | Yes | ||
| 62 | SHC3 | 4722 | 1.132 | 0.2478 | Yes | ||
| 63 | CLIC1 | 4725 | 1.131 | 0.2527 | Yes | ||
| 64 | BNIP3 | 4730 | 1.129 | 0.2575 | Yes | ||
| 65 | TNXB | 4821 | 1.112 | 0.2575 | Yes | ||
| 66 | DIRAS1 | 5514 | 0.980 | 0.2243 | No | ||
| 67 | NDRG2 | 5515 | 0.980 | 0.2286 | No | ||
| 68 | GKN1 | 5531 | 0.977 | 0.2321 | No | ||
| 69 | PRX | 5710 | 0.946 | 0.2267 | No | ||
| 70 | ABCD1 | 5769 | 0.935 | 0.2277 | No | ||
| 71 | VDR | 5930 | 0.903 | 0.2230 | No | ||
| 72 | GADD45A | 5991 | 0.894 | 0.2237 | No | ||
| 73 | SLC38A3 | 6001 | 0.893 | 0.2271 | No | ||
| 74 | GDNF | 6389 | 0.822 | 0.2098 | No | ||
| 75 | PRDM1 | 6917 | 0.726 | 0.1844 | No | ||
| 76 | HOXA11 | 6953 | 0.721 | 0.1857 | No | ||
| 77 | NRAS | 7112 | 0.696 | 0.1802 | No | ||
| 78 | CRYBA2 | 7134 | 0.692 | 0.1821 | No | ||
| 79 | FSTL1 | 7175 | 0.684 | 0.1829 | No | ||
| 80 | LRRTM3 | 7198 | 0.680 | 0.1848 | No | ||
| 81 | USP3 | 7263 | 0.669 | 0.1842 | No | ||
| 82 | CMYA1 | 7264 | 0.669 | 0.1872 | No | ||
| 83 | BACH1 | 7275 | 0.667 | 0.1896 | No | ||
| 84 | EN1 | 7399 | 0.645 | 0.1858 | No | ||
| 85 | AKT3 | 7452 | 0.635 | 0.1858 | No | ||
| 86 | TLL1 | 7712 | 0.584 | 0.1743 | No | ||
| 87 | SYNGR1 | 7723 | 0.582 | 0.1763 | No | ||
| 88 | IBRDC2 | 7731 | 0.580 | 0.1785 | No | ||
| 89 | NTN4 | 7735 | 0.579 | 0.1809 | No | ||
| 90 | MYBPH | 7797 | 0.567 | 0.1801 | No | ||
| 91 | SLC26A1 | 7813 | 0.565 | 0.1818 | No | ||
| 92 | COBL | 7824 | 0.563 | 0.1837 | No | ||
| 93 | CAMKK1 | 7923 | 0.543 | 0.1808 | No | ||
| 94 | ANXA7 | 7991 | 0.532 | 0.1796 | No | ||
| 95 | AK5 | 7998 | 0.531 | 0.1816 | No | ||
| 96 | PKP3 | 8278 | 0.478 | 0.1685 | No | ||
| 97 | DDIT3 | 8294 | 0.473 | 0.1698 | No | ||
| 98 | FOSL1 | 8432 | 0.449 | 0.1644 | No | ||
| 99 | MMP9 | 8581 | 0.419 | 0.1582 | No | ||
| 100 | SH3RF2 | 8713 | 0.397 | 0.1528 | No | ||
| 101 | MAPRE3 | 8799 | 0.382 | 0.1499 | No | ||
| 102 | PVALB | 8871 | 0.369 | 0.1477 | No | ||
| 103 | MAMDC2 | 9029 | 0.338 | 0.1407 | No | ||
| 104 | COL7A1 | 9085 | 0.326 | 0.1391 | No | ||
| 105 | PPP1R9B | 9478 | 0.238 | 0.1189 | No | ||
| 106 | RBP4 | 9660 | 0.197 | 0.1100 | No | ||
| 107 | BTK | 9728 | 0.183 | 0.1071 | No | ||
| 108 | ESRRG | 9810 | 0.163 | 0.1034 | No | ||
| 109 | FABP4 | 9845 | 0.158 | 0.1023 | No | ||
| 110 | LMOD3 | 9865 | 0.155 | 0.1020 | No | ||
| 111 | CMAS | 9920 | 0.144 | 0.0997 | No | ||
| 112 | SYT2 | 10005 | 0.127 | 0.0957 | No | ||
| 113 | IGSF9 | 10017 | 0.125 | 0.0956 | No | ||
| 114 | AP2M1 | 10074 | 0.114 | 0.0931 | No | ||
| 115 | PLCD1 | 10085 | 0.112 | 0.0930 | No | ||
| 116 | BLMH | 10129 | 0.102 | 0.0912 | No | ||
| 117 | LOR | 10215 | 0.082 | 0.0869 | No | ||
| 118 | GJA1 | 10216 | 0.082 | 0.0873 | No | ||
| 119 | CHST1 | 10247 | 0.076 | 0.0860 | No | ||
| 120 | SLC24A4 | 10279 | 0.070 | 0.0846 | No | ||
| 121 | EDG1 | 10290 | 0.068 | 0.0844 | No | ||
| 122 | PHLDA2 | 10313 | 0.061 | 0.0834 | No | ||
| 123 | ABCF3 | 10634 | -0.009 | 0.0661 | No | ||
| 124 | RGS2 | 10672 | -0.020 | 0.0642 | No | ||
| 125 | WNT7B | 10716 | -0.031 | 0.0620 | No | ||
| 126 | FBXW11 | 10879 | -0.066 | 0.0535 | No | ||
| 127 | FBXO2 | 11062 | -0.107 | 0.0441 | No | ||
| 128 | PTPRN | 11119 | -0.119 | 0.0416 | No | ||
| 129 | FBS1 | 11189 | -0.137 | 0.0384 | No | ||
| 130 | FGF11 | 11573 | -0.222 | 0.0186 | No | ||
| 131 | SYTL1 | 11741 | -0.262 | 0.0107 | No | ||
| 132 | RELA | 11860 | -0.290 | 0.0056 | No | ||
| 133 | NR0B2 | 12027 | -0.331 | -0.0019 | No | ||
| 134 | TRAF3 | 12121 | -0.351 | -0.0054 | No | ||
| 135 | MYH14 | 12241 | -0.380 | -0.0102 | No | ||
| 136 | DHRS3 | 12292 | -0.394 | -0.0112 | No | ||
| 137 | ABHD4 | 12430 | -0.427 | -0.0167 | No | ||
| 138 | ADAM15 | 12553 | -0.462 | -0.0213 | No | ||
| 139 | LAMC1 | 12575 | -0.465 | -0.0204 | No | ||
| 140 | CALB2 | 12654 | -0.484 | -0.0225 | No | ||
| 141 | HCN3 | 12688 | -0.493 | -0.0221 | No | ||
| 142 | GNAI1 | 12720 | -0.500 | -0.0216 | No | ||
| 143 | BAZ2A | 12744 | -0.508 | -0.0206 | No | ||
| 144 | CDKN1A | 12803 | -0.522 | -0.0214 | No | ||
| 145 | IL11 | 12922 | -0.551 | -0.0254 | No | ||
| 146 | SLC16A6 | 12954 | -0.561 | -0.0246 | No | ||
| 147 | WNT6 | 13085 | -0.599 | -0.0290 | No | ||
| 148 | SNX10 | 13098 | -0.603 | -0.0270 | No | ||
| 149 | SSH2 | 13194 | -0.627 | -0.0293 | No | ||
| 150 | DYRK1A | 13236 | -0.637 | -0.0288 | No | ||
| 151 | BCL9L | 13268 | -0.649 | -0.0276 | No | ||
| 152 | GTPBP2 | 13528 | -0.726 | -0.0384 | No | ||
| 153 | KCNA2 | 13630 | -0.762 | -0.0405 | No | ||
| 154 | SNCB | 13695 | -0.778 | -0.0406 | No | ||
| 155 | UCN2 | 13786 | -0.807 | -0.0419 | No | ||
| 156 | OMG | 13867 | -0.827 | -0.0426 | No | ||
| 157 | RIN1 | 14128 | -0.912 | -0.0526 | No | ||
| 158 | DTNA | 14437 | -1.008 | -0.0649 | No | ||
| 159 | HCLS1 | 14443 | -1.009 | -0.0607 | No | ||
| 160 | GRIA1 | 14478 | -1.020 | -0.0580 | No | ||
| 161 | PSMD11 | 14541 | -1.037 | -0.0568 | No | ||
| 162 | CRYGS | 15110 | -1.253 | -0.0821 | No | ||
| 163 | NEFH | 15266 | -1.313 | -0.0847 | No | ||
| 164 | LAMA3 | 15394 | -1.361 | -0.0856 | No | ||
| 165 | FGF12 | 15404 | -1.365 | -0.0800 | No | ||
| 166 | TMEM24 | 15437 | -1.378 | -0.0757 | No | ||
| 167 | XPOT | 15510 | -1.408 | -0.0733 | No | ||
| 168 | MYB | 15624 | -1.455 | -0.0730 | No | ||
| 169 | EMP1 | 16104 | -1.676 | -0.0916 | No | ||
| 170 | TOB1 | 16156 | -1.695 | -0.0869 | No | ||
| 171 | TAF15 | 16176 | -1.702 | -0.0804 | No | ||
| 172 | ATP1B1 | 16399 | -1.812 | -0.0844 | No | ||
| 173 | PSME4 | 16440 | -1.828 | -0.0785 | No | ||
| 174 | TIAL1 | 16584 | -1.898 | -0.0779 | No | ||
| 175 | BLR1 | 16618 | -1.921 | -0.0712 | No | ||
| 176 | RNF34 | 16782 | -2.008 | -0.0711 | No | ||
| 177 | RAB3D | 16896 | -2.065 | -0.0681 | No | ||
| 178 | XLKD1 | 17446 | -2.379 | -0.0874 | No | ||
| 179 | PSMD4 | 17477 | -2.395 | -0.0785 | No | ||
| 180 | SFRS2 | 17548 | -2.439 | -0.0715 | No | ||
| 181 | LAMC2 | 17616 | -2.484 | -0.0641 | No | ||
| 182 | H3F3B | 17752 | -2.572 | -0.0601 | No | ||
| 183 | TTC1 | 17892 | -2.678 | -0.0558 | No | ||
| 184 | VAPA | 17910 | -2.692 | -0.0448 | No | ||
| 185 | RBBP7 | 17920 | -2.697 | -0.0333 | No | ||
| 186 | PPP2CA | 18211 | -2.984 | -0.0359 | No | ||
| 187 | SRPK2 | 18286 | -3.115 | -0.0261 | No | ||
| 188 | CNTF | 18293 | -3.120 | -0.0127 | No | ||
| 189 | UBE2E3 | 18359 | -3.230 | -0.0019 | No | ||
| 190 | GTF2B | 18495 | -3.569 | 0.0066 | No |
