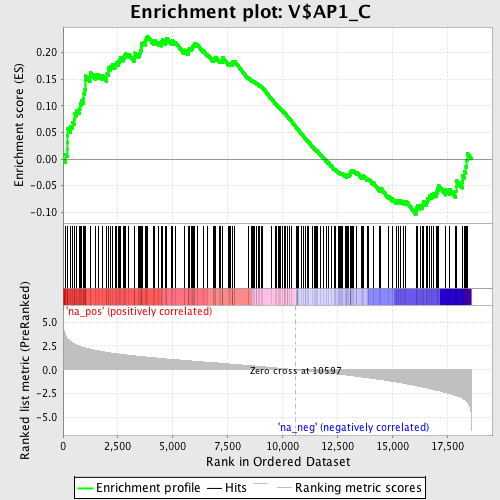
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$AP1_C |
| Enrichment Score (ES) | 0.23091044 |
| Normalized Enrichment Score (NES) | 1.1992842 |
| Nominal p-value | 0.09072165 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AIG1 | 111 | 3.556 | 0.0086 | Yes | ||
| 2 | ETV5 | 182 | 3.305 | 0.0183 | Yes | ||
| 3 | POLD4 | 185 | 3.299 | 0.0318 | Yes | ||
| 4 | IL10 | 195 | 3.274 | 0.0447 | Yes | ||
| 5 | TYSND1 | 215 | 3.223 | 0.0569 | Yes | ||
| 6 | MARK4 | 352 | 2.944 | 0.0616 | Yes | ||
| 7 | RB1CC1 | 421 | 2.822 | 0.0695 | Yes | ||
| 8 | VASP | 522 | 2.698 | 0.0751 | Yes | ||
| 9 | ROM1 | 534 | 2.685 | 0.0855 | Yes | ||
| 10 | MNT | 624 | 2.589 | 0.0913 | Yes | ||
| 11 | CLDN15 | 739 | 2.477 | 0.0953 | Yes | ||
| 12 | CDH23 | 770 | 2.460 | 0.1038 | Yes | ||
| 13 | RHBDF1 | 844 | 2.396 | 0.1096 | Yes | ||
| 14 | SLC35B1 | 930 | 2.330 | 0.1146 | Yes | ||
| 15 | SLITRK6 | 948 | 2.320 | 0.1232 | Yes | ||
| 16 | PLEC1 | 993 | 2.286 | 0.1302 | Yes | ||
| 17 | TUBB4 | 1007 | 2.279 | 0.1388 | Yes | ||
| 18 | CSF3 | 1008 | 2.276 | 0.1481 | Yes | ||
| 19 | LMNA | 1032 | 2.264 | 0.1562 | Yes | ||
| 20 | SEMA3B | 1228 | 2.148 | 0.1544 | Yes | ||
| 21 | HSPB7 | 1232 | 2.147 | 0.1631 | Yes | ||
| 22 | TBC1D17 | 1481 | 2.032 | 0.1579 | Yes | ||
| 23 | HEPH | 1614 | 1.977 | 0.1589 | Yes | ||
| 24 | CD68 | 1788 | 1.910 | 0.1573 | Yes | ||
| 25 | ADRA1A | 1996 | 1.825 | 0.1535 | Yes | ||
| 26 | AQP5 | 1997 | 1.824 | 0.1610 | Yes | ||
| 27 | IVL | 2081 | 1.795 | 0.1639 | Yes | ||
| 28 | USP2 | 2084 | 1.793 | 0.1711 | Yes | ||
| 29 | CHST4 | 2158 | 1.766 | 0.1744 | Yes | ||
| 30 | PEA15 | 2235 | 1.738 | 0.1774 | Yes | ||
| 31 | ITPKC | 2365 | 1.697 | 0.1774 | Yes | ||
| 32 | TRAPPC3 | 2416 | 1.683 | 0.1816 | Yes | ||
| 33 | CSMD3 | 2517 | 1.658 | 0.1829 | Yes | ||
| 34 | TCF7 | 2566 | 1.644 | 0.1871 | Yes | ||
| 35 | MPP5 | 2631 | 1.627 | 0.1903 | Yes | ||
| 36 | RICS | 2746 | 1.592 | 0.1906 | Yes | ||
| 37 | ANK3 | 2794 | 1.579 | 0.1945 | Yes | ||
| 38 | VIT | 2844 | 1.565 | 0.1983 | Yes | ||
| 39 | SEC24D | 2987 | 1.525 | 0.1968 | Yes | ||
| 40 | FIGF | 3253 | 1.454 | 0.1884 | Yes | ||
| 41 | HSPB8 | 3271 | 1.450 | 0.1934 | Yes | ||
| 42 | RPS6KA4 | 3274 | 1.450 | 0.1993 | Yes | ||
| 43 | FBXO44 | 3427 | 1.412 | 0.1968 | Yes | ||
| 44 | MYOZ2 | 3500 | 1.398 | 0.1986 | Yes | ||
| 45 | SMPX | 3518 | 1.394 | 0.2034 | Yes | ||
| 46 | MDFI | 3553 | 1.386 | 0.2073 | Yes | ||
| 47 | MAP1A | 3556 | 1.386 | 0.2129 | Yes | ||
| 48 | EPHB2 | 3570 | 1.382 | 0.2178 | Yes | ||
| 49 | WDFY3 | 3640 | 1.369 | 0.2197 | Yes | ||
| 50 | RIT1 | 3744 | 1.341 | 0.2196 | Yes | ||
| 51 | HDAC3 | 3770 | 1.334 | 0.2237 | Yes | ||
| 52 | SV2B | 3782 | 1.332 | 0.2286 | Yes | ||
| 53 | ADCK4 | 3840 | 1.321 | 0.2309 | Yes | ||
| 54 | COL16A1 | 4132 | 1.260 | 0.2203 | No | ||
| 55 | CLSTN3 | 4182 | 1.250 | 0.2227 | No | ||
| 56 | SFN | 4343 | 1.213 | 0.2190 | No | ||
| 57 | ELAVL2 | 4470 | 1.184 | 0.2170 | No | ||
| 58 | MARK1 | 4488 | 1.180 | 0.2210 | No | ||
| 59 | LAPTM5 | 4515 | 1.174 | 0.2244 | No | ||
| 60 | LTBP3 | 4682 | 1.140 | 0.2200 | No | ||
| 61 | SHC3 | 4722 | 1.132 | 0.2226 | No | ||
| 62 | CLIC1 | 4725 | 1.131 | 0.2271 | No | ||
| 63 | STAT5B | 4945 | 1.090 | 0.2197 | No | ||
| 64 | CAPN6 | 4965 | 1.087 | 0.2231 | No | ||
| 65 | GABARAPL1 | 5118 | 1.057 | 0.2192 | No | ||
| 66 | DIRAS1 | 5514 | 0.980 | 0.2017 | No | ||
| 67 | GKN1 | 5531 | 0.977 | 0.2049 | No | ||
| 68 | KBTBD5 | 5699 | 0.947 | 0.1997 | No | ||
| 69 | PRX | 5710 | 0.946 | 0.2030 | No | ||
| 70 | FGF9 | 5732 | 0.942 | 0.2057 | No | ||
| 71 | ABCD1 | 5769 | 0.935 | 0.2076 | No | ||
| 72 | DTX2 | 5831 | 0.922 | 0.2081 | No | ||
| 73 | NUDT11 | 5883 | 0.914 | 0.2091 | No | ||
| 74 | SCEL | 5913 | 0.907 | 0.2112 | No | ||
| 75 | VDR | 5930 | 0.903 | 0.2141 | No | ||
| 76 | GADD45A | 5991 | 0.894 | 0.2145 | No | ||
| 77 | SLC38A3 | 6001 | 0.893 | 0.2176 | No | ||
| 78 | IRAK1 | 6121 | 0.869 | 0.2147 | No | ||
| 79 | GDNF | 6389 | 0.822 | 0.2036 | No | ||
| 80 | PTPRR | 6595 | 0.784 | 0.1957 | No | ||
| 81 | NOTCH4 | 6838 | 0.737 | 0.1856 | No | ||
| 82 | CPNE8 | 6843 | 0.736 | 0.1884 | No | ||
| 83 | PRDM1 | 6917 | 0.726 | 0.1874 | No | ||
| 84 | LPP | 6929 | 0.723 | 0.1898 | No | ||
| 85 | HOXA11 | 6953 | 0.721 | 0.1915 | No | ||
| 86 | CRYBA2 | 7134 | 0.692 | 0.1845 | No | ||
| 87 | FSTL1 | 7175 | 0.684 | 0.1852 | No | ||
| 88 | TRPV3 | 7249 | 0.672 | 0.1839 | No | ||
| 89 | USP3 | 7263 | 0.669 | 0.1860 | No | ||
| 90 | CMYA1 | 7264 | 0.669 | 0.1887 | No | ||
| 91 | BACH1 | 7275 | 0.667 | 0.1909 | No | ||
| 92 | VAT1 | 7556 | 0.613 | 0.1782 | No | ||
| 93 | CLDN17 | 7575 | 0.612 | 0.1798 | No | ||
| 94 | IL9 | 7629 | 0.600 | 0.1793 | No | ||
| 95 | TLL1 | 7712 | 0.584 | 0.1773 | No | ||
| 96 | SYNGR1 | 7723 | 0.582 | 0.1791 | No | ||
| 97 | IBRDC2 | 7731 | 0.580 | 0.1811 | No | ||
| 98 | NTN4 | 7735 | 0.579 | 0.1834 | No | ||
| 99 | MYBPH | 7797 | 0.567 | 0.1824 | No | ||
| 100 | SLC26A1 | 7813 | 0.565 | 0.1839 | No | ||
| 101 | FOSL1 | 8432 | 0.449 | 0.1521 | No | ||
| 102 | MET | 8434 | 0.448 | 0.1539 | No | ||
| 103 | MMP9 | 8581 | 0.419 | 0.1477 | No | ||
| 104 | PCDH9 | 8611 | 0.414 | 0.1478 | No | ||
| 105 | SLCO5A1 | 8678 | 0.402 | 0.1459 | No | ||
| 106 | SH3RF2 | 8713 | 0.397 | 0.1457 | No | ||
| 107 | MAPRE3 | 8799 | 0.382 | 0.1426 | No | ||
| 108 | SFTPC | 8834 | 0.376 | 0.1423 | No | ||
| 109 | EPHA2 | 8891 | 0.366 | 0.1408 | No | ||
| 110 | XYLT1 | 8972 | 0.350 | 0.1379 | No | ||
| 111 | MAMDC2 | 9029 | 0.338 | 0.1362 | No | ||
| 112 | COL7A1 | 9085 | 0.326 | 0.1346 | No | ||
| 113 | PPP1R9B | 9478 | 0.238 | 0.1142 | No | ||
| 114 | RBP4 | 9660 | 0.197 | 0.1052 | No | ||
| 115 | BTK | 9728 | 0.183 | 0.1023 | No | ||
| 116 | ESRRG | 9810 | 0.163 | 0.0986 | No | ||
| 117 | FABP4 | 9845 | 0.158 | 0.0974 | No | ||
| 118 | CMAS | 9920 | 0.144 | 0.0940 | No | ||
| 119 | SYT2 | 10005 | 0.127 | 0.0899 | No | ||
| 120 | AP2M1 | 10074 | 0.114 | 0.0867 | No | ||
| 121 | PLCD1 | 10085 | 0.112 | 0.0866 | No | ||
| 122 | BLMH | 10129 | 0.102 | 0.0847 | No | ||
| 123 | LOR | 10215 | 0.082 | 0.0804 | No | ||
| 124 | GJA1 | 10216 | 0.082 | 0.0808 | No | ||
| 125 | PHLDA2 | 10313 | 0.061 | 0.0758 | No | ||
| 126 | EFNA1 | 10420 | 0.037 | 0.0702 | No | ||
| 127 | MCF2 | 10624 | -0.007 | 0.0592 | No | ||
| 128 | ABCF3 | 10634 | -0.009 | 0.0587 | No | ||
| 129 | RGS2 | 10672 | -0.020 | 0.0568 | No | ||
| 130 | PROCR | 10677 | -0.021 | 0.0567 | No | ||
| 131 | SPRED1 | 10692 | -0.025 | 0.0560 | No | ||
| 132 | WNT7B | 10716 | -0.031 | 0.0549 | No | ||
| 133 | FBXW11 | 10879 | -0.066 | 0.0464 | No | ||
| 134 | BTG3 | 10941 | -0.082 | 0.0434 | No | ||
| 135 | GADD45G | 11047 | -0.104 | 0.0381 | No | ||
| 136 | FBXO2 | 11062 | -0.107 | 0.0378 | No | ||
| 137 | SERPINB5 | 11141 | -0.123 | 0.0341 | No | ||
| 138 | FBS1 | 11189 | -0.137 | 0.0321 | No | ||
| 139 | USP13 | 11347 | -0.171 | 0.0242 | No | ||
| 140 | ASB5 | 11450 | -0.200 | 0.0195 | No | ||
| 141 | DSCR1L1 | 11489 | -0.207 | 0.0183 | No | ||
| 142 | ANKRD22 | 11561 | -0.221 | 0.0154 | No | ||
| 143 | FGF11 | 11573 | -0.222 | 0.0157 | No | ||
| 144 | SYTL1 | 11741 | -0.262 | 0.0077 | No | ||
| 145 | RELA | 11860 | -0.290 | 0.0025 | No | ||
| 146 | S100A5 | 11986 | -0.321 | -0.0030 | No | ||
| 147 | ULK1 | 12079 | -0.343 | -0.0066 | No | ||
| 148 | PITPNC1 | 12209 | -0.371 | -0.0121 | No | ||
| 149 | FLNC | 12368 | -0.412 | -0.0190 | No | ||
| 150 | ABHD4 | 12430 | -0.427 | -0.0206 | No | ||
| 151 | ADAM15 | 12553 | -0.462 | -0.0253 | No | ||
| 152 | LAMC1 | 12575 | -0.465 | -0.0245 | No | ||
| 153 | CALB2 | 12654 | -0.484 | -0.0268 | No | ||
| 154 | HCN3 | 12688 | -0.493 | -0.0265 | No | ||
| 155 | BAZ2A | 12744 | -0.508 | -0.0274 | No | ||
| 156 | ZFYVE9 | 12847 | -0.532 | -0.0308 | No | ||
| 157 | POLR3E | 12916 | -0.550 | -0.0322 | No | ||
| 158 | IL11 | 12922 | -0.551 | -0.0302 | No | ||
| 159 | SLC16A6 | 12954 | -0.561 | -0.0296 | No | ||
| 160 | PKN3 | 13001 | -0.575 | -0.0298 | No | ||
| 161 | NFRKB | 13075 | -0.596 | -0.0313 | No | ||
| 162 | LENEP | 13077 | -0.597 | -0.0289 | No | ||
| 163 | WNT6 | 13085 | -0.599 | -0.0268 | No | ||
| 164 | SNX10 | 13098 | -0.603 | -0.0250 | No | ||
| 165 | VCL | 13115 | -0.607 | -0.0234 | No | ||
| 166 | PDGFRB | 13139 | -0.613 | -0.0221 | No | ||
| 167 | SSH2 | 13194 | -0.627 | -0.0225 | No | ||
| 168 | DYRK1A | 13236 | -0.637 | -0.0221 | No | ||
| 169 | CENTD1 | 13377 | -0.685 | -0.0269 | No | ||
| 170 | EYA1 | 13385 | -0.687 | -0.0244 | No | ||
| 171 | APBA2BP | 13620 | -0.758 | -0.0340 | No | ||
| 172 | CAPN12 | 13662 | -0.769 | -0.0331 | No | ||
| 173 | SNCB | 13695 | -0.778 | -0.0317 | No | ||
| 174 | OMG | 13867 | -0.827 | -0.0376 | No | ||
| 175 | EPHA1 | 13935 | -0.845 | -0.0377 | No | ||
| 176 | RIN1 | 14128 | -0.912 | -0.0444 | No | ||
| 177 | DTNA | 14437 | -1.008 | -0.0570 | No | ||
| 178 | GRIA1 | 14478 | -1.020 | -0.0550 | No | ||
| 179 | SLC22A18 | 14843 | -1.149 | -0.0701 | No | ||
| 180 | PACSIN3 | 15016 | -1.213 | -0.0744 | No | ||
| 181 | ATP6V1C2 | 15184 | -1.280 | -0.0782 | No | ||
| 182 | NEFH | 15266 | -1.313 | -0.0773 | No | ||
| 183 | LAMA3 | 15394 | -1.361 | -0.0786 | No | ||
| 184 | XPOT | 15510 | -1.408 | -0.0790 | No | ||
| 185 | MYB | 15624 | -1.455 | -0.0792 | No | ||
| 186 | IDS | 16086 | -1.665 | -0.0974 | No | ||
| 187 | EMP1 | 16104 | -1.676 | -0.0915 | No | ||
| 188 | TOB1 | 16156 | -1.695 | -0.0873 | No | ||
| 189 | SNCG | 16281 | -1.751 | -0.0868 | No | ||
| 190 | ATP1B1 | 16399 | -1.812 | -0.0857 | No | ||
| 191 | PSME4 | 16440 | -1.828 | -0.0804 | No | ||
| 192 | TIAL1 | 16584 | -1.898 | -0.0804 | No | ||
| 193 | BLR1 | 16618 | -1.921 | -0.0743 | No | ||
| 194 | ATP6V1B2 | 16676 | -1.944 | -0.0694 | No | ||
| 195 | RNF34 | 16782 | -2.008 | -0.0669 | No | ||
| 196 | RAB3D | 16896 | -2.065 | -0.0646 | No | ||
| 197 | TLX2 | 17009 | -2.124 | -0.0619 | No | ||
| 198 | PPP2CB | 17059 | -2.149 | -0.0558 | No | ||
| 199 | MCART1 | 17125 | -2.188 | -0.0503 | No | ||
| 200 | XLKD1 | 17446 | -2.379 | -0.0579 | No | ||
| 201 | LAMC2 | 17616 | -2.484 | -0.0569 | No | ||
| 202 | TTC1 | 17892 | -2.678 | -0.0609 | No | ||
| 203 | VAPA | 17910 | -2.692 | -0.0507 | No | ||
| 204 | RBBP7 | 17920 | -2.697 | -0.0402 | No | ||
| 205 | SDCBP | 18186 | -2.956 | -0.0424 | No | ||
| 206 | PPP2CA | 18211 | -2.984 | -0.0315 | No | ||
| 207 | CNTF | 18293 | -3.120 | -0.0231 | No | ||
| 208 | UBE2E3 | 18359 | -3.230 | -0.0134 | No | ||
| 209 | ROCK2 | 18399 | -3.300 | -0.0019 | No | ||
| 210 | LRRFIP2 | 18413 | -3.333 | 0.0110 | No |
